Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae
- PMID: 15037778
- PMCID: PMC1262634
- DOI: 10.1261/rna.5147804
Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae
Abstract
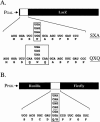
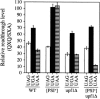
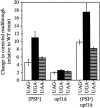
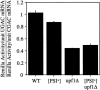
The Nonsense-Mediated mRNA Decay (NMD) pathway mediates the rapid degradation of mRNAs that contain premature stop mutations in eukaryotic organisms. It was recently shown that mutations in three yeast genes that encode proteins involved in the NMD process, UPF1, UPF2, and UPF3, also reduce the efficiency of translation termination. In the current study, we compared the efficiency of translation termination in a upf1Delta strain and a [PSI(+)] strain using a collection of translation termination reporter constructs. The [PSI(+)] state is caused by a prion form of the polypeptide chain release factor eRF3 that limits its availability to participate in translation termination. In contrast, the mechanism by which Upf1p influences translation termination is poorly understood. The efficiency of translation termination is primarily determined by a tetranucleotide termination signal consisting of the stop codon and the first nucleotide immediately 3' of the stop codon. We found that the upf1Delta mutation, like the [PSI(+)] state, decreases the efficiency of translation termination over a broad range of tetranucleotide termination signals in a unique, context-dependent manner. These results suggest that Upf1p may associate with the termination complex prior to polypeptide chain release. We also found that the increase in readthrough observed in a [PSI(+)]/upf1Delta strain was larger than the readthrough observed in strains carrying either defect alone, indicating that the upf1Delta mutation and the [PSI(+)] state influence the termination process in distinct ways. Finally, our analysis revealed that the mRNA destabilization associated with NMD could be separated into two distinct forms that correlated with the extent the premature stop codon was suppressed. The minor component of NMD was a 25% decrease in mRNA levels observed when readthrough was >/=0.5%, while the major component was represented by a larger decrease in mRNA abundance that was observed only when readthrough was </=0.5%. This low threshold for the onset of the major component of NMD indicates that mRNA surveillance is an ongoing process that occurs throughout the lifetime of an mRNA.
Figures

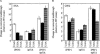

Similar articles
-
Inactivation of NMD increases viability of sup45 nonsense mutants in Saccharomyces cerevisiae.BMC Mol Biol. 2007 Aug 16;8:71. doi: 10.1186/1471-2199-8-71. BMC Mol Biol. 2007. PMID: 17705828 Free PMC article.
-
ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons.Nat Commun. 2016 Dec 23;7:14021. doi: 10.1038/ncomms14021. Nat Commun. 2016. PMID: 28008922 Free PMC article.
-
Nonsense-mediated mRNA decay factors cure most [PSI+] prion variants.Proc Natl Acad Sci U S A. 2018 Feb 6;115(6):E1184-E1193. doi: 10.1073/pnas.1717495115. Epub 2018 Jan 22. Proc Natl Acad Sci U S A. 2018. PMID: 29358398 Free PMC article.
-
Modulation of efficiency of translation termination in Saccharomyces cerevisiae.Prion. 2014;8(3):247-60. doi: 10.4161/pri.29851. Epub 2014 Nov 1. Prion. 2014. PMID: 25486049 Free PMC article. Review.
-
NMD monitors translational fidelity 24/7.Curr Genet. 2017 Dec;63(6):1007-1010. doi: 10.1007/s00294-017-0709-4. Epub 2017 May 23. Curr Genet. 2017. PMID: 28536849 Free PMC article. Review.
Cited by
-
Chemical-Induced Read-Through at Premature Termination Codons Determined by a Rapid Dual-Fluorescence System Based on S. cerevisiae.PLoS One. 2016 Apr 27;11(4):e0154260. doi: 10.1371/journal.pone.0154260. eCollection 2016. PLoS One. 2016. PMID: 27119736 Free PMC article.
-
GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination.Mol Cell Biol. 2004 Sep;24(17):7769-78. doi: 10.1128/MCB.24.17.7769-7778.2004. Mol Cell Biol. 2004. PMID: 15314182 Free PMC article.
-
A kinase-dependent checkpoint prevents escape of immature ribosomes into the translating pool.PLoS Biol. 2019 Dec 13;17(12):e3000329. doi: 10.1371/journal.pbio.3000329. eCollection 2019 Dec. PLoS Biol. 2019. Update in: PLoS Biol. 2024 Apr 25;22(4):e3001767. doi: 10.1371/journal.pbio.3001767. PMID: 31834877 Free PMC article. Updated.
-
Yeast Sup35 Prion Structure: Two Types, Four Parts, Many Variants.Int J Mol Sci. 2019 May 29;20(11):2633. doi: 10.3390/ijms20112633. Int J Mol Sci. 2019. PMID: 31146333 Free PMC article.
-
Global analysis of alternative splicing uncovers developmental regulation of nonsense-mediated decay in C. elegans.RNA. 2009 Sep;15(9):1652-60. doi: 10.1261/rna.1711109. Epub 2009 Jul 17. RNA. 2009. PMID: 19617316 Free PMC article.
References
-
- Adams, A., Gottschling, D.E., Kaiser, C.A., and Stearns, T. 1997. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Atkin, A.L., Schenkman, L.R., Eastham, M., Dahlseid, J.N., Lelivelt, M.J., and Culbertson, M.R. 1997. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem. 272: 22163–22172. - PubMed
-
- Beelman, C.A., Stevens, A., Caponigro, G., LaGrandeur, T.E., Hatfield, L., Fortner, D.M., and Parker, R. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646. - PubMed
-
- Bertram, G., Innes, S., Minella, O., Richardson, J., and Stansfield, I. 2001. Endless possibilities: Translation termination and stop codon recognition. Microbiology 147: 255–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
