Rat pro-inflammatory cytokine and cytokine related mRNA quantification by real-time polymerase chain reaction using SYBR green
- PMID: 15040812
- PMCID: PMC373448
- DOI: 10.1186/1471-2172-5-3
Rat pro-inflammatory cytokine and cytokine related mRNA quantification by real-time polymerase chain reaction using SYBR green
Abstract
Background: Cytokine mRNA quantification is widely used to investigate cytokine profiles, particularly in small samples. Real-time polymerase chain reaction is currently the most reliable method of quantifying low-level transcripts such as cytokine and cytokine receptor mRNAs. This accurate technique allows the quantification of a larger pattern of cytokines than quantification at the protein level, which is limited to a smaller number of proteins.
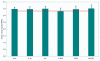
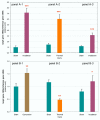
Results: Although fluorogenic probes are considered more sensitive than fluorescent dyes, we have developed SYBR Green real-time RT-PCR protocols to assay pro-inflammatory cytokines (IL1a, IL1b and IL6, TNFa), cytokine receptors (IL1-r1, IL1-r2, IL6-r, TNF-r2) and related molecules (IL1-RA, SOCS3) mRNA in rats. This method enables normalisation against several housekeeping genes (beta-actin, GAPDH, CypA, HPRT) dependent on the specific experimental treatments and tissues using either standard curve, or comparative CT quantification method. PCR efficiency and sensitivity allow the assessment of; i) basal mRNA levels in many tissues and even decreases in mRNA levels, ii) mRNA levels from very small samples.
Conclusion: Real-time RT-PCR is currently the best way to investigate cytokine networks. The investigations should be completed by the analysis of genes regulated by cytokines or involved in cytokine signalling, providing indirect information on cytokine protein expression.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

