Genomic structure and alternative splicing of murine R2B receptor protein tyrosine phosphatases (PTPkappa, mu, rho and PCP-2)
- PMID: 15040814
- PMCID: PMC373446
- DOI: 10.1186/1471-2164-5-14
Genomic structure and alternative splicing of murine R2B receptor protein tyrosine phosphatases (PTPkappa, mu, rho and PCP-2)
Abstract
Background: Four genes designated as PTPRK (PTPkappa), PTPRL/U (PCP-2), PTPRM (PTPmu) and PTPRT (PTPrho) code for a subfamily (type R2B) of receptor protein tyrosine phosphatases (RPTPs) uniquely characterized by the presence of an N-terminal MAM domain. These transmembrane molecules have been implicated in homophilic cell adhesion. In the human, the PTPRK gene is located on chromosome 6, PTPRL/U on 1, PTPRM on 18 and PTPRT on 20. In the mouse, the four genes ptprk, ptprl, ptprm and ptprt are located in syntenic regions of chromosomes 10, 4, 17 and 2, respectively.
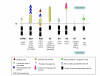
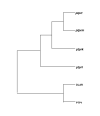
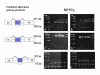
Results: The genomic organization of murine R2B RPTP genes is described. The four genes varied greatly in size ranging from approximately 64 kb to approximately 1 Mb, primarily due to proportional differences in intron lengths. Although there were also minor variations in exon length, the number of exons and the phases of exon/intron junctions were highly conserved. In situ hybridization with digoxigenin-labeled cRNA probes was used to localize each of the four R2B transcripts to specific cell types within the murine central nervous system. Phylogenetic analysis of complete sequences indicated that PTPrho and PTPmu were most closely related, followed by PTPkappa. The most distant family member was PCP-2. Alignment of RPTP polypeptide sequences predicted putative alternatively spliced exons. PCR experiments revealed that five of these exons were alternatively spliced, and that each of the four phosphatases incorporated them differently. The greatest variability in genomic organization and the majority of alternatively spliced exons were observed in the juxtamembrane domain, a region critical for the regulation of signal transduction.
Conclusions: Comparison of the four R2B RPTP genes revealed virtually identical principles of genomic organization, despite great disparities in gene size due to variations in intron length. Although subtle differences in exon length were also observed, it is likely that functional differences among these genes arise from the specific combinations of exons generated by alternative splicing.
Figures








References
-
- Schindelholz B, Knirr M, Warrior R, Zinn K. Regulation of CNS and motor axon guidance in Drosophila by the receptor tyrosine phosphatase DPTP52F. Development. 2001;128:4371–82. - PubMed
-
- Johnson KG, Van Vactor D. Receptor protein tyrosine phosphatases in nervous system development. Physiol Rev. 2003;83:1–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

