Dynamic water networks in cytochrome C oxidase from Paracoccus denitrificans investigated by molecular dynamics simulations
- PMID: 15041635
- PMCID: PMC1304046
- DOI: 10.1016/S0006-3495(04)74254-X
Dynamic water networks in cytochrome C oxidase from Paracoccus denitrificans investigated by molecular dynamics simulations
Abstract
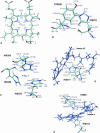
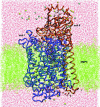
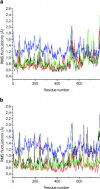
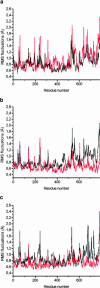
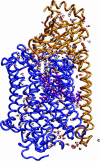
We present a molecular dynamics study of cytochrome c oxidase from Paracoccus denitrificans in the fully oxidized state, embedded in a fully hydrated dimyristoylphosphatidylcholine lipid bilayer membrane. Parallel simulations with different levels of protein hydration, 1.125 ns each in length, were carried out under conditions of constant temperature and pressure using three-dimensional periodic boundary conditions and full electrostatics to investigate the distribution and dynamics of water molecules and their corresponding hydrogen-bonded networks inside cytochrome c oxidase. The majority of the water molecules had residence times shorter than 100 ps, but a few water molecules are fixed inside the protein for up to 1.125 ns. The hydrogen-bonded network in cytochrome c oxidase is not uniformly distributed, and the degree of water arrangement is variable. The average number of solvent sites in the proton-conducting K- and D-pathways was determined. In contrast to single water files in narrow geometries we observe significant diffusion of individual water molecules along these pathways. The highly fluctuating hydrogen-bonded networks, combined with the significant diffusion of individual water molecules, provide a basis for the transfer of protons in cytochrome c oxidase, therefore leading to a better understanding of the mechanism of proton pumping.
Figures

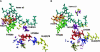

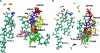
References
-
- Aagaard, A., G. Gilderson, D. A. Mills, S. Ferguson-Miller, and P. Brzezinski. 2000. Redesign of the proton-pumping machinery of cytochrome c oxidase: proton pumping does not require Glu(I-286). Biochemistry. 39:15847–15850. - PubMed
-
- Babcock, G. T., and M. Wikström. 1992. Oxygen activation and the conservation of energy in cell respiration. Nature. 356:301–309. - PubMed
-
- Backgren, C., G. Hummer, M. Wikström, and A. Puustinen. 2000. Proton translocation by cytochrome c oxidase can take place without the conserved glutamic acid in subunit I. Biochemistry. 39:7863–7867. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

