HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: possible contributions to drug resistance and a potential new target site for drugs
- PMID: 15044738
- PMCID: PMC2280056
- DOI: 10.1110/ps.03468904
HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: possible contributions to drug resistance and a potential new target site for drugs
Erratum in
- Protein Sci. 2004 May;13(5):1434
Abstract
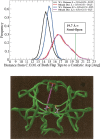
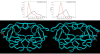
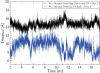
The protease from type 1 human immunodeficiency virus (HIV-1) is a critical drug target against which many therapeutically useful inhibitors have been developed; however, the set of viral strains in the population has been shifting to become more drug-resistant. Because indirect effects are contributing to drug resistance, an examination of the dynamic structures of a wild-type and a mutant could be insightful. Consequently, this study examined structural properties sampled during 22 nsec, all atom molecular dynamics (MD) simulations (in explicit water) of both a wild-type and the drug-resistant V82F/I84V mutant of HIV-1 protease. The V82F/I84V mutation significantly decreases the binding affinity of all HIV-1 protease inhibitors currently used clinically. Simulations have shown that the curling of the tips of the active site flaps immediately results in flap opening. In the 22-nsec MD simulations presented here, more frequent and more rapid curling of the mutant's active site flap tips was observed. The mutant protease's flaps also opened farther than the wild-type's flaps did and displayed more flexibility. This suggests that the effect of the mutations on the equilibrium between the semiopen and closed conformations could be one aspect of the mechanism of drug resistance for this mutant. In addition, correlated fluctuations in the active site and periphery were noted that point to a possible binding site for allosteric inhibitors.
Figures














References
-
- Ala, P.J., Huston, E.E., Klabe, R.M., McCabe, D.D., Duke, J.L., Rizzo, C.J., Korant, B.D., DeLoskey, R.J., Lam, P.Y.S., Hodge, C.N., et al. 1997. Molecular basis of HIV-1 protease drug resistance: Structural analysis of mutant proteases complexed with cyclic urea inhibitors. Biochemistry 36 1573–1580. - PubMed
-
- Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, W.F., DiNola, A., and Haak, J.R. 1984. Molecular dynamics with coupling to an external bath. J. Chem.Phys. 81 3684–3690.
-
- Case, D.A., Pearlman, D.A., Caldwell, J.W., Cheatham III, T.E., Wang J., Ross, W.S., Simmerling, C., Darden, T., Merz, K.M., Stanton, R.V., et al. 2002. AMBER 7. University of California, San Francisco.
-
- Collins, J.R., Burt, S.K., and Erickson, J.W. 1995. Flap opening in HIV-1 protease simulated by “activated” molecular dynamics. Nat. Struct. Biol. 2 334–338. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

