Solution structure of a novel calcium binding protein, MTH1880, from Methanobacterium thermoautotrophicum
- PMID: 15044740
- PMCID: PMC2280053
- DOI: 10.1110/ps.03472104
Solution structure of a novel calcium binding protein, MTH1880, from Methanobacterium thermoautotrophicum
Abstract
MTH1880 is a hypothetical protein from Methanobacterium thermoautotrophicum, a target organism of structural genomics. The solution structure determined by NMR spectroscopy demonstrates a typical alpha + beta-fold found in many proteins with different functions. The molecular surface of the protein reveals a small, highly acidic pocket comprising loop B (Asp36, Asp37, Asp38), the end of beta2 (Glu39), and loop D (Ser57, Ser58, Ser61), indicating that the protein would have a possible cation binding site. The NMR resonances of several amino acids within the acidic binding pocket in MTH1880, shifted upon addition of calcium ion. This calcium binding motif and overall topology of MTH1880 differ from those of other calcium binding proteins. MTH1880 did not show a calcium-induced conformational change typical of calcium sensor proteins. Therefore, we propose that the MTH1880 protein contains a novel motif for calcium-specific binding, and may function as a calcium buffering protein.
Figures


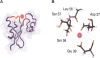
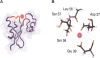
References
-
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kun-stleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 54 905–921. - PubMed
-
- Christendat, D., Yee, A., Dharamsi, A., Kluger, Y., Savchenko, A., Cort, J.R., Booth, V., Mackereth, C.D., Saridakis, V., Ekiel, I., et al. 2000. Structural proteomics of an archaeon. Nat. Struct. Biol. 7 903–909. - PubMed
-
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. - PubMed
-
- Goddard, T.D and Kneller, D.G. 2003. SPARKY 3. University of California, San Francisco, CA.
-
- Holm, L. and Sander, C. 1995. Dali: A network tool for protein structure comparison. Trends Biochem. Sci. 20 478–480. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

