Dissection and design of yeast prions
- PMID: 15045026
- PMCID: PMC374241
- DOI: 10.1371/journal.pbio.0020086
Dissection and design of yeast prions
Abstract
Many proteins can misfold into beta-sheet-rich, self-seeding polymers (amyloids). Prions are exceptional among such aggregates in that they are also infectious. In fungi, prions are not pathogenic but rather act as epigenetic regulators of cell physiology, providing a powerful model for studying the mechanism of prion replication. We used prion-forming domains from two budding yeast proteins (Sup35p and New1p) to examine the requirements for prion formation and inheritance. In both proteins, a glutamine/asparagine-rich (Q/N-rich) tract mediates sequence-specific aggregation, while an adjacent motif, the oligopeptide repeat, is required for the replication and stable inheritance of these aggregates. Our findings help to explain why although Q/N-rich proteins are relatively common, few form heritable aggregates: prion inheritance requires both an aggregation sequence responsible for self-seeded growth and an element that permits chaperone-dependent replication of the aggregate. Using this knowledge, we have designed novel artificial prions by fusing the replication element of Sup35p to aggregation-prone sequences from other proteins, including pathogenically expanded polyglutamine.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures

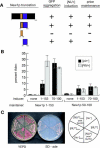

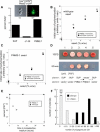


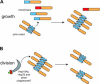
Comment in
-
Yeast prions: protein aggregation is not enough.PLoS Biol. 2004 Apr;2(4):E125. doi: 10.1371/journal.pbio.0020125. Epub 2004 Apr 13. PLoS Biol. 2004. PMID: 15094820 Free PMC article.
References
-
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. - PubMed
-
- Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP, et al. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol. 2000;35:865–876. - PubMed
-
- Chien P, Weissman JS. Conformational diversity in a yeast prion dictates its seeding specificity. Nature. 2001;410:223–227. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

