Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity
- PMID: 15047530
- PMCID: PMC375300
- DOI: 10.1128/AAC.48.4.1281-1288.2004
Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity
Abstract
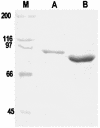
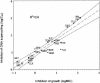
Genome studies suggest that DNA gyrase is the sole type II topoisomerase and likely the unique target of quinolones in Mycobacterium tuberculosis. Despite the emerging importance of quinolones in the treatment of mycobacterial disease, the slow growth and high pathogenicity of M. tuberculosis have precluded direct purification of its gyrase and detailed analysis of quinolone action. To address these issues, we separately overexpressed the M. tuberculosis DNA gyrase GyrA and GyrB subunits as His-tagged proteins in Escherichia coli from pET plasmids carrying gyrA and gyrB genes. The soluble 97-kDa GyrA and 72-kDa GyrB subunits were purified by nickel chelate chromatography and shown to reconstitute an ATP-dependent DNA supercoiling activity. The drug concentration that inhibited DNA supercoiling by 50% (IC(50)) was measured for 22 different quinolones, and values ranged from 2 to 3 microg/ml (sparfloxacin, sitafloxacin, clinafloxacin, and gatifloxacin) to >1,000 microg/ml (pipemidic acid and nalidixic acid). By comparison, MICs measured against M. tuberculosis ranged from 0.12 microg/ml (for gatifloxacin) to 128 microg/ml (both pipemidic acid and nalidixic acid) and correlated well with the gyrase IC(50)s (R(2) = 0.9). Quinolones promoted gyrase-mediated cleavage of plasmid pBR322 DNA due to stabilization of the cleavage complex, which is thought to be the lethal lesion. Surprisingly, the measured concentrations of drug inducing 50% plasmid linearization correlated less well with the MICs (R(2) = 0.7). These findings suggest that the DNA supercoiling inhibition assay may be a useful screening test in identifying quinolones with promising activity against M. tuberculosis. The quinolone structure-activity relationship demonstrated here shows that C-8, the C-7 ring, the C-6 fluorine, and the N-1 cyclopropyl substituents are desirable structural features in targeting M. tuberculosis gyrase.
Figures

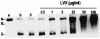


References
-
- Alovero, F. L., X. S. Pan, J. E. Morris, R. H. Manzo, and L. M. Fisher. 2000. Engineering the specificity of antibacterial fluoroquinolones: benzenesulfonamide modifications at C-7 of ciprofloxacin change its primary target in Streptococcus pneumoniae from topoisomerase IV to gyrase. Antimicrob. Agents Chemother. 44:320-325. - PMC - PubMed
-
- Banerjee, S. K., K. Bhatt, S. Rana, P. Misra, and P. K. Chakraborti. 1996. Involvement of an efflux system in mediating high level of fluoroquinolone resistance in Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 226:362-368. - PubMed
-
- Barrett, J. F., J. I. Bernstein, H. M. Krause, J. J. Hilliard, and K. A. Ohemeng. 1993. Testing potential gyrase inhibitors of bacterial DNA gyrase: a comparison of the supercoiling inhibition assay and “cleavable complex” assay. Anal. Biochem. 214:313-317. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

