tRNA-like structure regulates translation of Brome mosaic virus RNA
- PMID: 15047816
- PMCID: PMC374274
- DOI: 10.1128/jvi.78.8.4003-4010.2004
tRNA-like structure regulates translation of Brome mosaic virus RNA
Abstract
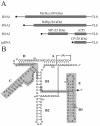
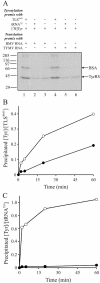
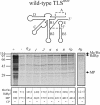
For various groups of plant viruses, the genomic RNAs end with a tRNA-like structure (TLS) instead of the 3' poly(A) tail of common mRNAs. The actual function of these TLSs has long been enigmatic. Recently, however, it became clear that for turnip yellow mosaic virus, a tymovirus, the valylated TLS(TYMV) of the single genomic RNA functions as a bait for host ribosomes and directs them to the internal initiation site of translation (with N-terminal valine) of the second open reading frame for the polyprotein. This discovery prompted us to investigate whether the much larger TLSs of a different genus of viruses have a comparable function in translation. Brome mosaic virus (BMV), a bromovirus, has a tripartite RNA genome with a subgenomic RNA4 for coat protein expression. All four RNAs carry a highly conserved and bulky 3' TLS(BMV) (about 200 nucleotides) with determinants for tyrosylation. We discovered TLS(BMV)-catalyzed self-tyrosylation of the tyrosyl-tRNA synthetase but could not clearly detect tyrosine incorporation into any virus-encoded protein. We established that BMV proteins do not need TLS(BMV) tyrosylation for their initiation. However, disruption of the TLSs strongly reduced the translation of genomic RNA1, RNA2, and less strongly, RNA3, whereas coat protein expression from RNA4 remained unaffected. This aberrant translation could be partially restored by providing the TLS(BMV) in trans. Intriguingly, a subdomain of the TLS(BMV) could even almost fully restore translation to the original pattern. We discuss here a model with a central and dominant role for the TLS(BMV) during the BMV infection cycle.
Figures


References
-
- Ahlquist, P., R. Dasgupta, and P. Kaesberg. 1981. Near identity of 3′ RNA secondary structure in bromoviruses and cucumber mosaic virus. Cell 23:183-189. - PubMed
-
- Barends, S., H. H. J. Bink, S. H. E. van den Worm, C. W. A. Pleij, and B. Kraal. 2003. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell 112:123-129. - PubMed
-
- Bol, J. F. 2003. Alfalfa mosaic virus: coat protein-dependent initiation of infection. Mol. Plant Pathol. 4:1-8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

