The differential sensitivity of human and rhesus macaque CCR5 to small-molecule inhibitors of human immunodeficiency virus type 1 entry is explained by a single amino acid difference and suggests a mechanism of action for these inhibitors
- PMID: 15047829
- PMCID: PMC374253
- DOI: 10.1128/jvi.78.8.4134-4144.2004
The differential sensitivity of human and rhesus macaque CCR5 to small-molecule inhibitors of human immunodeficiency virus type 1 entry is explained by a single amino acid difference and suggests a mechanism of action for these inhibitors
Abstract
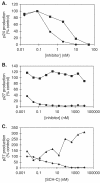
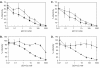
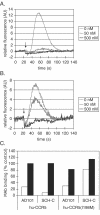
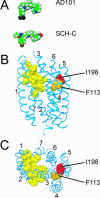
AD101 and SCH-C are two chemically related small molecules that inhibit the entry of human immunodeficiency virus type 1 (HIV-1) via human CCR5. AD101 also inhibits HIV-1 entry via rhesus macaque CCR5, but SCH-C does not. Among the eight residues that differ between the human and macaque versions of the coreceptor, only one, methionine-198, accounts for the insensitivity of macaque CCR5 to inhibition by SCH-C. Thus, the macaque coreceptor engineered to contain the natural human CCR5 residue (isoleucine) at position 198 is sensitive to HIV-1 entry inhibition by SCH-C, whereas a human CCR5 mutant containing the corresponding macaque residue (methionine) is resistant. Position 198 is in CCR5 transmembrane (TM) helix 5 and is not located within the previously defined binding site for AD101 and SCH-C, which involves residues in TM helices 1, 2, 3, and 7. SCH-C binds to human CCR5 whether residue 198 is isoleucine or methionine, and it also binds to macaque CCR5. However, the binding of a conformation-dependent monoclonal antibody to human CCR5 is inhibited by SCH-C only when residue 198 is isoleucine. These observations, taken together, suggest that the antiviral effects of SCH-C and AD101 involve stabilization, or induction, of a CCR5 conformation that is not compatible with HIV-1 infection. However, SCH-C is unable to exert this effect on CCR5 conformation when residue 198 is methionine. The region of CCR5 near residue 198 has, therefore, an important influence on the conformational state of this receptor.
Figures
References
-
- Ansari-Lari, M. A., X. M. Liu, M. L. Metzker, A. R. Rut, and R. A. Gibbs. 1997. The extent of genetic variation in the CCR5 gene. Nat. Genet. 16:221-222. - PubMed
-
- Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 96:5698-5703. - PMC - PubMed
-
- Blanpain, C., B. J. Doranz, A. Bondue, C. Govaerts, A. De Leener, G. Vassart, R. W. Doms, A. Proudfoot, and M. Parmentier. 2003. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J. Biol. Chem. 278:5179-5187. - PubMed
-
- Blanpain, C., B. Lee, M. Tackoen, B. Puffer, A. Boom, F. Libert, M. Sharron, V. Wittamer, G. Vassart, R. W. Doms, and M. Parmentier. 2000. Multiple nonfunctional alleles of CCR5 are frequent in various human populations. Blood 96:1638-1645. - PubMed
-
- Carrington, M., M. Dean, M. P. Martin, and S. J. O'Brien. 1999. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum. Mol. Genet. 8:1939-1945. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

