Role of minority populations of human immunodeficiency virus type 1 in the evolution of viral resistance to protease inhibitors
- PMID: 15047838
- PMCID: PMC374292
- DOI: 10.1128/jvi.78.8.4234-4247.2004
Role of minority populations of human immunodeficiency virus type 1 in the evolution of viral resistance to protease inhibitors
Abstract
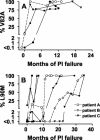
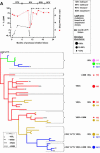
Human immunodeficiency virus type 1 (HIV-1) drug resistance results from the accumulation of mutations in the viral genes targeted by the drugs. These genetic changes, however, are commonly detected and monitored by techniques that only take into account the dominant population of plasma virus. Because HIV-1-infected patients harbor a complex and diverse mixture of virus populations, the mechanisms underlying the emergence and the evolution of resistance are not fully elucidated. Using techniques that allow the quantification of resistance mutations in minority virus species, we have monitored the evolution of resistance in plasma virus populations from patients failing protease inhibitor treatment. Minority populations with distinct resistance genotypes were detected in all patients throughout the evolution of resistance. The emergence of new dominant genotypes followed two possible mechanisms: (i) emergence of a new mutation in a currently dominant genotype and (ii) emergence of a new genotype derived from a minority virus species. In most cases, these population changes were associated with an increase in resistance at the expense of a reduction in replication capacity. Our findings provide a preliminary indication that minority viral species, which evolve independently of the majority virus population, can eventually become dominant populations, thereby serving as a reservoir of diversity and possibly accelerating the development of drug resistance.
Figures





References
-
- Archer, R. H., C. Dykes, P. Gerondelis, A. Lloyd, P. Fay, R. C. Reichman, R. A. Bambara, and L. M. Demeter. 2000. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J. Virol. 74:8390-8401. - PMC - PubMed
-
- Barbosa, P., P. Charneau, N. Dumey, and F. Clavel. 1994. Kinetic analysis of HIV-1 early replicative steps in a coculture system. AIDS Res. Hum. Retrovir. 10:53-59. - PubMed
-
- Barbour, J. D., T. Wrin, R. M. Grant, J. N. Martin, M. R. Segal, C. J. Petropoulos, and S. G. Deeks. 2002. Evolution of phenotypic drug susceptibility and viral replication capacity during long-term virologic failure of protease inhibitor therapy in human immunodeficiency virus-infected adults. J. Virol. 76:11104-11112. - PMC - PubMed
-
- Bleiber, G., M. Munoz, A. Ciuffi, P. Meylan, and A. Telenti. 2001. Individual contributions of mutant protease and reverse transcriptase to viral infectivity, replication, and protein maturation of antiretroviral drug-resistant human immunodeficiency virus type 1. J. Virol. 75:3291-3300. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

