Non-random positioning of chromosomes in human sperm nuclei
- PMID: 15053486
- PMCID: PMC1409707
- DOI: 10.1023/b:chro.0000013166.04629.97
Non-random positioning of chromosomes in human sperm nuclei
Abstract
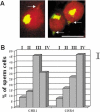
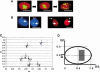
In human spermatozoa, the arrangement of chromosomes is non-random. Characteristic features are association of centromeres in the interior chromocenter and peripheral location of telomeres. In this paper, we have investigated the highest level of order in DNA packing in sperm--absolute and relative intranuclear chromosome positioning. Asymmetrical nuclear shape, existence of a defined spatial marker, and the haploid complement of chromosomes facilitated an experimental approach using in situ hybridization. Our results showed the tendency for non-random intranuclear location of individual chromosome territories. Moreover, centromeres demonstrated specific intranuclear position, and were located within a limited area of nuclear volume. Additionally, the relative positions of centromeres were non-random; some were found in close proximity, while other pairs showed significantly greater intercentromere distances. Therefore, a unique and specific adherence may exist between chromosomes in sperm. The observed chromosome order is discussed in relation to sperm nuclei decondensation, and reactivation during fertilization.
Figures


References
-
- Ashley T. Specific end-to-end attachment of chromosomes in Ornithogalum virens. J Cell Sci. 1979;38:357–367. - PubMed
-
- Bickmore WA, Chubb JR. Chromosome position: now, where was I? Curr Biol. 2003;13:357–359. - PubMed
-
- Bojanowski K, Maniotis AJ, Plisov S, Larsen AK, Ingber DE. DNA topoisomerase II can drive changes in higher order chromosome architecture without enzymatically modifying DNA. J Cell Biochem. 1998;69:127–142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
