Prediction of Saccharomyces cerevisiae replication origins
- PMID: 15059255
- PMCID: PMC395781
- DOI: 10.1186/gb-2004-5-4-r22
Prediction of Saccharomyces cerevisiae replication origins
Abstract
Background: Autonomously replicating sequences (ARSs) function as replication origins in Saccharomyces cerevisiae. ARSs contain the 17 bp ARS consensus sequence (ACS), which binds the origin recognition complex. The yeast genome contains more than 10,000 ACS matches, but there are only a few hundred origins, and little flanking sequence similarity has been found. Thus, identification of origins by sequence alone has not been possible.
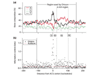
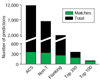
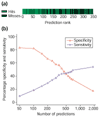
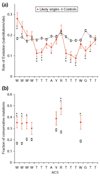
Results: We developed an algorithm, Oriscan, to predict yeast origins using similarity to 26 characterized origins. Oriscan used 268 bp of sequence, including the T-rich ACS and a 3' A-rich region. The predictions identified the exact location of the ACS. A total of 84 of the top 100 Oriscan predictions, and 56% of the top 350, matched known ARSs or replication protein binding sites. The true accuracy was even higher because we tested 25 discrepancies, and 15 were in fact ARSs. Thus, 94% of the top 100 predictions and an estimated 70% of the top 350 were correct. We compared the predictions to corresponding sequences in related Saccharomyces species and found that the ACSs of experimentally supported predictions show significant conservation.
Conclusions: The high accuracy of the predictions indicates that we have defined near-sufficient conditions for ARS activity, the A-rich region is a recognizable feature of ARS elements with a probable role in replication initiation, and nucleotide sequence is a reliable predictor of yeast origins. Oriscan detected most origins in the genome, demonstrating previously unrecognized generality in yeast replication origins and significant discriminatory power in the algorithm.
Figures

References
-
- Jacob F, Brenner S, Cuzin F. On the regulation of DNA replication in bacteria. Cold Spring Harb Symp Quant Biol. 1963;28:329–438.
-
- Newlon CS, Collins I, Dershowitz A, Deshpande AM, Greenfeder SA, Ong LY, Theis JF. Analysis of replication origin function on chromosome III of Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1993;58:415–423. - PubMed
-
- Stinchcomb DT, Struhl K, Davis RW. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979;282:39–43. - PubMed
-
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. - PubMed
-
- Huberman JA, Spotila LD, Nawotka KA, el-Assouli SM, Davis LR. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987;51:473–481. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

