A transcriptional timetable of autumn senescence
- PMID: 15059257
- PMCID: PMC395783
- DOI: 10.1186/gb-2004-5-4-r24
A transcriptional timetable of autumn senescence
Abstract
Background: We have developed genomic tools to allow the genus Populus (aspens and cottonwoods) to be exploited as a full-featured model for investigating fundamental aspects of tree biology. We have undertaken large-scale expressed sequence tag (EST) sequencing programs and created Populus microarrays with significant gene coverage. One of the important aspects of plant biology that cannot be studied in annual plants is the gene activity involved in the induction of autumn leaf senescence.
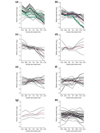
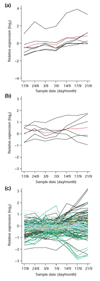
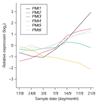
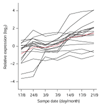
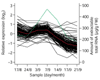
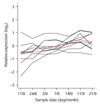
Results: On the basis of 36,354 Populus ESTs, obtained from seven cDNA libraries, we have created a DNA microarray consisting of 13,490 clones, spotted in duplicate. Of these clones, 12,376 (92%) were confirmed by resequencing and all sequences were annotated and functionally classified. Here we have used the microarray to study transcript abundance in leaves of a free-growing aspen tree (Populus tremula) in northern Sweden during natural autumn senescence. Of the 13,490 spotted clones, 3,792 represented genes with significant expression in all leaf samples from the seven studied dates.
Conclusions: We observed a major shift in gene expression, coinciding with massive chlorophyll degradation, that reflected a shift from photosynthetic competence to energy generation by mitochondrial respiration, oxidation of fatty acids and nutrient mobilization. Autumn senescence had much in common with senescence in annual plants; for example many proteases were induced. We also found evidence for increased transcriptional activity before the appearance of visible signs of senescence, presumably preparing the leaf for degradation of its components.
Figures





References
-
- The virtual library of plant-arrays: Publications http://www.univ-montp2.fr/~plant_arrays/Plant-Arrays_Publications.html
-
- Sterky F, Regan S, Karlsson J, Hertzberg M, Rohde A, Holmberg A, Amini B, Bhalerao R, Larsson M, Villarroel R, et al. Gene discovery in the wood-forming tissues of poplar: analysis of 5,692 expressed sequence tags. Proc Natl Acad Sci USA. 1998;95:13330–13335. doi: 10.1073/pnas.95.22.13330. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

