Comparative analysis of amino acid repeats in rodents and humans
- PMID: 15059995
- PMCID: PMC383298
- DOI: 10.1101/gr.1925704
Comparative analysis of amino acid repeats in rodents and humans
Abstract
Amino acid tandem repeats, also called homopolymeric tracts, are extremely abundant in eukaryotic proteins. To gain insight into the genome-wide evolution of these regions in mammals, we analyzed the repeat content in a large data set of rat-mouse-human orthologs. Our results show that human proteins contain more amino acid repeats than rodent proteins and that trinucleotide repeats are also more abundant in human coding sequences. Using the human species as an outgroup, we were able to address differences in repeat loss and repeat gain in the rat and mouse lineages. In this data set, mouse proteins contain substantially more repeats than rat proteins, which can be at least partly attributed to a higher repeat loss in the rat lineage. The data are consistent with a role for trinucleotide slippage in the generation of novel amino acid repeats. We confirm the previously observed functional bias of proteins with repeats, with overrepresentation of transcription factors and DNA-binding proteins. We show that genes encoding amino acid repeats tend to have an unusually high GC content, and that differences in coding GC content among orthologs are directly related to the presence/absence of repeats. We propose that the different GC content isochore structure in rodents and humans may result in an increased amino acid repeat prevalence in the human lineage.
Figures
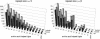

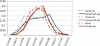

References
-
- Adkins, R.M., Gelke, E.L., Rowe, D., and Honeycutt, R.L. 2001. Molecular phylogeny and divergence time estimates for major rodent groups: Evidence from multiple genes. Mol. Biol. Evol. 18: 777-791. - PubMed
-
- Albà, M.M., Santibáñez-Koref, M.F., and Hancock, J.M. 1999a. Amino acid reiterations in yeast are over-represented in particular classes of proteins and show evidence of a slippage-like mutational process. J. Mol. Evol. 49: 789-797. - PubMed
-
- Albà, M.M., Santibáñez-Koref, M.F., and Hancock, J.M. 1999b. Conservation of polyglutamine tract size between mouse and human depends on codon interruption. Mol. Biol. Evol. 16: 1641-1644. - PubMed
-
- Albà, M.M., Santibáñez-Koref, M.F., and Hancock, J.M. 2001. The comparative genomics of polyglutamine repeats: Extreme difference in the codon organization of repeat-encoding regions between mammals and Drosophila. J. Mol. Evol. 52: 249-259. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
