The genomic sequence and comparative analysis of the rat major histocompatibility complex
- PMID: 15060004
- PMCID: PMC383307
- DOI: 10.1101/gr.1987704
The genomic sequence and comparative analysis of the rat major histocompatibility complex
Abstract
We have determined the sequence of a 4-Mb interval on rat chromosome 20p12 that encompasses the rat major histocompatibility complex (MHC). This is the first report of a finished sequence for a segment of the rat genome and constitutes one of the largest contiguous sequences thus far for rodent genomes in general. The rat MHC is, next to the human MHC, the second mammalian MHC sequenced to completion. Our analysis has resulted in the identification of at least 220 genes located within the sequenced interval. Although gene content and order are well conserved in the class II and class III gene intervals as well as the framework gene regions, profound rat-specific features were encountered within the class I gene regions, in comparison to human and mouse. Class I region-associated differences were found both at the structural level, the number, and organization of class I genes and gene families, and, in a more global context, in the way that evolution worked to shape the present-day rat MHC.
Figures

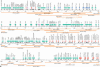

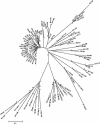

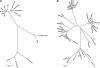
References
-
- Amadou, C. 1999. Evolution of the MHC class I region: The framework hypothesis. Immunogenetics 49: 362-367. - PubMed
-
- Anzai, T., Shiina, T., Kimura, N., Yanagiya, K., Kohara, S., Shigenari, A., Yamagata, T., Kulski, J.K., Naruse, T.K., Fujimori, Y., et al. 2003. Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc. Natl. Acad. Sci. 100: 7708-7713. - PMC - PubMed
-
- Chardon, P., Renard, C., Rogel-Gaillard, C., Cattolico, L., Duprat, S., Vaiman, M., and Renard, C. 2001. Sequence of the swine major histocompatibility complex region containing all nonclassical class I genes. Tissue Antigens 57: 55-65. - PubMed
-
- Druet, E., Sapin, C., Günther, E., Feingold, N., and Druet, P. 1977. Mercuric chloride-induced antiglomerular basement membrane antibodies in the rat: Genetic control. Eur. J. Immunol. 7: 348-351. - PubMed
-
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186-194. - PubMed
WEB SITE REFERENCES
-
- http://www.rzpd.de; German Resource Center.
-
- http://www.sanger.ac.uk/Software/sequencing/docs/phrap2gap; Sequence assembly software.
-
- http://www.ornl.gov/hgmis/research/bermuda.html#1; Policies on release of genomic sequence data.
-
- http://ncbi.nlm.nih.gov/LocusLink; LocusLink database. Provides a single query interface to curated sequence and descriptive information about genetic loci.
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
