Identification of a novel gene (HSN2) causing hereditary sensory and autonomic neuropathy type II through the Study of Canadian Genetic Isolates
- PMID: 15060842
- PMCID: PMC1181970
- DOI: 10.1086/420795
Identification of a novel gene (HSN2) causing hereditary sensory and autonomic neuropathy type II through the Study of Canadian Genetic Isolates
Abstract
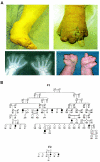
Hereditary sensory and autonomic neuropathy (HSAN) type II is an autosomal recessive disorder characterized by impairment of pain, temperature, and touch sensation owing to reduction or absence of peripheral sensory neurons. We identified two large pedigrees segregating the disorder in an isolated population living in Newfoundland and performed a 5-cM genome scan. Linkage analysis identified a locus mapping to 12p13.33 with a maximum LOD score of 8.4. Haplotype sharing defined a candidate interval of 1.06 Mb containing all or part of seven annotated genes, sequencing of which failed to detect causative mutations. Comparative genomics revealed a conserved ORF corresponding to a novel gene in which we found three different truncating mutations among five families including patients from rural Quebec and Nova Scotia. This gene, termed "HSN2," consists of a single exon located within intron 8 of the PRKWNK1 gene and is transcribed from the same strand. The HSN2 protein may play a role in the development and/or maintenance of peripheral sensory neurons or their supporting Schwann cells.
Figures


References
Electronic-Database Information
-
- Gene Discovery Program for Functional Genomics in Pig Reproduction, http://pigest.genome.iastate.edu/
-
- HUGO Gene Nomenclature Committee, http://www.gene.ucl.ac.uk/nomenclature/
References
-
- Basu S, Paul DK, Basu S (2002) Four siblings with type II hereditary sensory and autonomic neuropathy. Indian Pediatr 39:870–874 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

