Molecular analyses of the Arabidopsis TUBBY-like protein gene family
- PMID: 15064372
- PMCID: PMC419833
- DOI: 10.1104/pp.103.037820
Molecular analyses of the Arabidopsis TUBBY-like protein gene family
Abstract
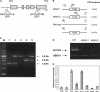
In mammals, TUBBY-like proteins play an important role in maintenance and function of neuronal cells during postdifferentiation and development. We have identified a TUBBY-like protein gene family with 11 members in Arabidopsis, named AtTLP1-11. Although seven of the AtTLP genes are located on chromosome I, no local tandem repeats or gene clusters are identified. Except for AtTLP4, reverse transcription-PCR analysis indicates that all these genes are expressed in various organs in 6-week-old Arabidopsis. AtTLP1, 2, 3, 6, 7, 9, 10, and 11 are expressed ubiquitously in all the organs tested, but the expression of AtTLP5 and 8 shows dramatic organ specificity. These 11 family members share 30% to 80% amino acid similarities across their conserved C-terminal tubby domains. Unlike the highly diverse N-terminal region of animal TUBBY-like proteins, all AtTLP members except AtTLP8 contain a conserved F-box domain (51-57 residues). The interaction between AtTLP9 and ASK1 (Arabidopsis Skp1-like 1) is confirmed via yeast (Saccharomyces cerevisiae) two-hybrid assays. Abscisic acid (ABA)-insensitive phenotypes are observed for two independent AtTLP9 mutant lines, whereas transgenic plants overexpressing AtTLP9 are hypersensitive to ABA. These results suggest that AtTLP9 may participate in the ABA signaling pathway.
Figures







References
-
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274 - PubMed
-
- Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro SL (1999) Implication of tubby proteins as transcription factors by structure-based functional analysis. Science 286: 2119–2125 - PubMed
-
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 - PubMed
-
- Coleman DL, Eicher EM (1990) Fat (fat) and tubby (tub): two autosomal recessive mutations causing obesity syndromes in the mouse. J Hered 81: 424–427 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

