WWOX binds the specific proline-rich ligand PPXY: identification of candidate interacting proteins
- PMID: 15064722
- PMCID: PMC4143251
- DOI: 10.1038/sj.onc.1207680
WWOX binds the specific proline-rich ligand PPXY: identification of candidate interacting proteins
Abstract
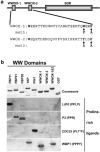
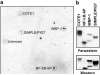
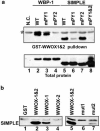
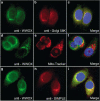
WWOX, the gene that maps to common chromosomal fragile site FRA16D, is frequently affected by aberrations in multiple types of cancers. WWOX encodes a 46 kDa protein that contains two WW domains and a short-chain oxidoreductase (SDR) domain. We recently demonstrated that ectopic expression of WWOX inhibits xenograft tumor growth of tumorigenic breast cancer cells. Little is known of the biochemical function(s) of WWOX. The SDR domain is predicted to be involved in sex-steroid metabolism and the WW domains are likely involved in protein-protein interactions. In this report, we identify the specific proline-rich ligand for WWOX as PPXY and show that the amino-terminal WW domain is responsible for this interaction. Using the WWOX WW domains as a probe, we screened high-density protein arrays and identified five candidate-binding partners. The binding to one of these candidates, small membrane protein of the lysosome/late endosome (SIMPLE), was further analysed, and we observed that a specific PPSY motif in the SIMPLE amino-acid sequence was required to interact with the amino-terminal WW domain of WWOX. In addition, immunofluorescence staining demonstrated that endogenous WWOX and SIMPLE co-localize to perinuclear compartments of MCF-7 human breast cancer cells. These studies demonstrate that WWOX contains a Group I WW domain that binds known cellular proteins containing the specific ligand PPXY. Identification and characterization of WWOX interacting proteins will lead to an understanding of the biological functions of WWOX in normal and tumor cells.
Figures
References
-
- Bedford MT, Sarbassova D, Xu J, Leder P, Yaffe MB. J. Biol. Chem. 2000;275:10359–10369. - PubMed
-
- Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ, Aldaz CM. Cancer Res. 2001;61:8068–8073. - PubMed
-
- Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. Cancer Res. 2000;60:2140–2145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

