Exposure of Saccharomyces cerevisiae to acetaldehyde induces sulfur amino acid metabolism and polyamine transporter genes, which depend on Met4p and Haa1p transcription factors, respectively
- PMID: 15066780
- PMCID: PMC383134
- DOI: 10.1128/AEM.70.4.1913-1922.2004
Exposure of Saccharomyces cerevisiae to acetaldehyde induces sulfur amino acid metabolism and polyamine transporter genes, which depend on Met4p and Haa1p transcription factors, respectively
Abstract
Acetaldehyde is a toxic compound produced by Saccharomyces cerevisiae cells under several growth conditions. The adverse effects of this molecule are important, as significant amounts accumulate inside the cells. By means of global gene expression analyses, we have detected the effects of acetaldehyde addition in the expression of about 400 genes. Repressed genes include many genes involved in cell cycle control, cell polarity, and the mitochondrial protein biosynthesis machinery. Increased expression is displayed in many stress response genes, as well as other families of genes, such as those encoding vitamin B1 biosynthesis machinery and proteins for aryl alcohol metabolism. The induction of genes involved in sulfur metabolism is dependent on Met4p and other well-known factors involved in the transcription of MET genes under nonrepressing conditions of sulfur metabolism. Moreover, the deletion of MET4 leads to increased acetaldehyde sensitivity. TPO genes encoding polyamine transporters are also induced by acetaldehyde; in this case, the regulation is dependent on the Haa1p transcription factor. In this paper, we discuss the connections between acetaldehyde and the processes affected by this compound in yeast cells with reference to the microarray data.
Figures

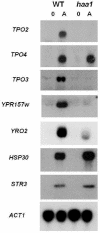


References
-
- Albertsen, M., I. Bellahn, R. Krämer, and S. Waffenschmidt. 2003. Localization and function of the yeast multidrug transporter Tpo1p. J. Biol. Chem. 278:12820-12825. - PubMed
-
- Alexandre, H., V. Ansanay-Galeote, S. Dequin, and B. Blondin. 2001. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 498:98-103. - PubMed
-
- Aranda, A., A. Querol, and M. del Olmo. 2002. Correlation between acetaldehyde and ethanol resistance and expression of HSP genes in yeast strains isolated during the biological aging of sherry wines. Arch. Microbiol. 177:304-312. - PubMed
-
- Aranda, A., and M. del Olmo. 2003. Response to acetaldehyde stress in the yeast Saccharomyces cerevisiae involves a strain-dependent regulation of several ALD genes and is mediated by the general stress response pathway. Yeast 20:749-759. - PubMed
-
- Backhus, L. E., J. DeRisi, P. O. Brown, and L. F. Bisson. 2001. Functional genomic analysis of a commercial wine strain of Saccharomyces cerevisiae under differing nitrogen conditions. FEMS Yeast Res. 1413:1-15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

