Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons
- PMID: 15066784
- PMCID: PMC383131
- DOI: 10.1128/AEM.70.4.1944-1955.2004
Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons
Abstract
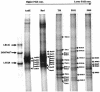
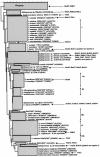
Bacterial strains of the genus Sphingomonas are often isolated from contaminated soils for their ability to use polycyclic aromatic hydrocarbons (PAH) as the sole source of carbon and energy. The direct detection of Sphingomonas strains in contaminated soils, either indigenous or inoculated, is, as such, of interest for bioremediation purposes. In this study, a culture-independent PCR-based detection method using specific primers targeting the Sphingomonas 16S rRNA gene combined with denaturing gradient gel electrophoresis (DGGE) was developed to assess Sphingomonas diversity in PAH-contaminated soils. PCR using the new primer pair on a set of template DNAs of different bacterial genera showed that the method was selective for bacteria belonging to the family Sphingomonadaceae.Single-band DGGE profiles were obtained for most Sphingomonas strains tested. Strains belonging to the same species had identical DGGE fingerprints, and in most cases, these fingerprints were typical for one species. Inoculated strains could be detected at a cell concentration of 10(4) CFU g of soil(-1). The analysis of Sphingomonas population structures of several PAH-contaminated soils by the new PCR-DGGE method revealed that soils containing the highest phenanthrene concentrations showed the lowest Sphingomonas diversity. Sequence analysis of cloned PCR products amplified from soil DNA revealed new 16S rRNA gene Sphingomonas sequences significantly different from sequences from known cultivated isolates (i.e., sequences from environmental clones grouped phylogenetically with other environmental clone sequences available on the web and that possibly originated from several potential new species). In conclusion, the newly designed Sphingomonas-specific PCR-DGGE detection technique successfully analyzed the Sphingomonas communities from polluted soils at the species level and revealed different Sphingomonas members not previously detected by culture-dependent detection techniques.
Figures

References
-
- Adkins, A. 1999. Degradation of the phenoxy acid herbicide diclofop-methyl by Sphingomonas paucimobilis isolated from a Canadian prairie soil. J. Ind. Microbiol. Biotechnol. 23:332-335. - PubMed
-
- Altschul, S., W. Gish, W. Miller, E. Myers, and D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Bastiaens, L. 1998. Ph.D. thesis. Catholic University of Leuven, Leuven, Belgium.
-
- Bastiaens, L., D. Springael, W. Dejonghe, P. Wattiau, H. Verachtert, and L. Diels. 2001. A transcriptional luxAB reporter fusion responding to fluorene in Sphingomonas sp. LB126 and its characterisation for whole-cell bioreporter purposes. Res. Microbiol. 152:849-859. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

