Mitochondrial DNA ligase in Crithidia fasciculata
- PMID: 15070723
- PMCID: PMC384752
- DOI: 10.1073/pnas.0305705101
Mitochondrial DNA ligase in Crithidia fasciculata
Abstract
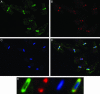
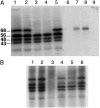
Kinetoplast DNA (kDNA), the form of mitochondrial DNA in trypanosomatids, consists of thousands of interlocked circular DNAs organized into a compact disk structure. A type II DNA topoisomerase, a DNA polymerase beta, and a structure-specific endonuclease have been localized to antipodal sites flanking the kDNA disk along with nascent DNA minicircles. We have cloned a gene (LIG k) encoding a mitochondrial DNA ligase in the trypanosomatid Crithidia fasciculata, and we show that an epitope-tagged form of the ligase colocalizes with the other replication proteins at the antipodal sites and also at the two faces of the kDNA disk. DNA LIG k becomes adenylated in reactions with ATP, and the adenylate moiety is removed by incubation with pyrophosphate or nicked DNA. The ligase interacts physically with the beta polymerase and is proposed to be involved in the repair of gaps in the newly synthesized minicircles. In yeast and mammals, a single gene encodes both nuclear and mitochondrial forms of DNA ligase. The LIG K protein sequence has low similarity to mitochondrial DNA ligases in other eukaryotes and is distinct from the C. fasciculata nuclear DNA ligase (LIG I).
Figures


Comment in
-
Closing the gaps in kinetoplast DNA network replication.Proc Natl Acad Sci U S A. 2004 Mar 30;101(13):4333-4. doi: 10.1073/pnas.0401400101. Epub 2004 Mar 22. Proc Natl Acad Sci U S A. 2004. PMID: 15070715 Free PMC article. Review. No abstract available.
References
-
- Shapiro, T. A. & Englund, P. T. (1995) in Annu. Rev. Microbiol., 49, 117–143. - PubMed
-
- Klingbeil, M. M., Drew, M. E., Liu, Y., Morris, J. C., Motyka, S. A., Saxowsky, T. T., Wang, Z. & Englund, P. T. (2001) Protist 152, 255–262. - PubMed
-
- Morris, J. C., Drew, M. E., Klingbeil, M. M., Motyka, S. A., Saxowsky, T. T., Wang, Z. & Englund, P. T. (2001) Int. J. Parasitol. 31, 453–458. - PubMed
-
- Sturm, N. R. & Simpson, L. (1990) Cell 61, 879–884. - PubMed
-
- Chen, J., Rauch, C. A., White, J. H., Englund, P. T. & Cozzarelli, N. R. (1995) Cell 80, 61–69. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

