Exploring amyloid formation by a de novo design
- PMID: 15070736
- PMCID: PMC384765
- DOI: 10.1073/pnas.0306786101
Exploring amyloid formation by a de novo design
Abstract
Protein deposition as amyloid fibrils underlies many debilitating human disorders. The complexity and size of disease-related polypeptides, however, often hinders a detailed rational approach to study effects that contribute to the process of amyloid formation. We report here a simplified peptide sequence successfully designed de novo to fold into a coiled-coil conformation under ambient conditions but to transform into amyloid fibrils at elevated temperatures. We have determined the crystal structure of the coiled-coil form and propose a detailed molecular model for the peptide in its fibrillar state. The relative stabilities of the two structural forms and the kinetics of their interconversion were found to be highly sensitive to small sequence changes. The results reveal the importance of specific packing interactions on the kinetics of amyloid formation and show the potential of this exceptionally favorable system for probing details of the molecular origins of amyloid disease.
Figures
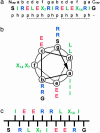
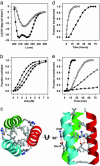
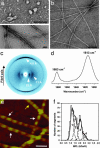
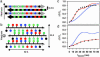
Comment in
-
Unzipping the mysteries of amyloid fiber formation.Proc Natl Acad Sci U S A. 2004 Mar 30;101(13):4335-6. doi: 10.1073/pnas.0401163101. Epub 2004 Mar 22. Proc Natl Acad Sci U S A. 2004. PMID: 15070716 Free PMC article. Review. No abstract available.
References
-
- Taylor, J. P., Hardy, J. & Fischbeck, K. H. (2002) Science 296, 1991–1995. - PubMed
-
- Hardy, J. & Selkoe, D. J. (2002) Science 297, 353–356. - PubMed
-
- Fändrich, M., Fletcher, M. A. & Dobson, C. M. (2001) Nature 410, 165–166. - PubMed
-
- Bucciantini, M., Giannoni, E., Chiti, F., Baroni, F., Formigli, L., Zurdo, J., Taddei, N., Ramponi, G., Dobson, C. M. & Stefani, M. (2002) Nature 416, 507–511. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

