WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis
- PMID: 15070782
- PMCID: PMC384811
- DOI: 10.1073/pnas.0305574101
WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis
Abstract
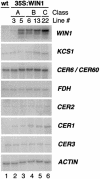
Epicuticular wax forms a layer of hydrophobic material on plant aerial organs, which constitutes a protective barrier between the plant and its environment. We report here the identification of WIN1, an Arabidopsis thaliana ethylene response factor-type transcription factor, which can activate wax deposition in overexpressing plants. We constitutively expressed WIN1 in transgenic Arabidopsis plants, and found that leaf epidermal wax accumulation was up to 4.5-fold higher in these plants than in control plants. A significant increase was also found in stems. Interestingly, approximately 50% of the additional wax could only be released by complete lipid extractions, suggesting that not all of the wax is superficial. Gene expression analysis indicated that a number of genes, such as CER1, KCS1, and CER2, which are known to be involved in wax biosynthesis, were induced in WIN1 overexpressors. This observation indicates that induction of wax accumulation in transgenic plants is probably mediated through an increase in the expression of genes encoding enzymes of the wax biosynthesis pathway.
Figures




References
-
- Riederer, M. & Schreiber, L. (2001) J. Exp. Bot. 52, 2023-2032. - PubMed
-
- Kerstiens, G. (1996) J. Exp. Bot. 47, 1813-1832.
-
- Eigenbrode, S. D. & Espelie, K. E. (1995) Annu. Rev. Entomol. 40, 171-194.
-
- Belding, R. D., Sutton, T. B., Blankenship, S. M. & Young, E. (2000) Plant Dis. 84, 767-772. - PubMed
-
- Marcell, L. M. & Beattie, G. A. (2002) Mol. Plant-Microbe Interact. 15, 1236-1244. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

