Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains
- PMID: 15071027
- PMCID: PMC387571
- DOI: 10.1128/JCM.42.4.1694-1702.2004
Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains
Abstract
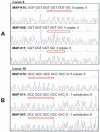
We describe a multilocus short sequence repeat (MLSSR) sequencing approach for the genotyping of Mycobacterium avium subsp. paratuberculosis (M. paratuberculosis) strains. Preliminary analysis identified 185 mono-, di-, and trinucleotide repeat sequences dispersed throughout the M. paratuberculosis genome, of which 78 were perfect repeats. Comparative nucleotide sequencing of the 78 loci of six M. paratuberculosis isolates from different host species and geographic locations identified a subset of 11 polymorphic short sequence repeats (SSRs), with an average of 3.2 alleles per locus. Comparative sequencing of these 11 loci was used to genotype a collection of 33 M. paratuberculosis isolates representing different multiplex PCR for IS900 loci (MPIL) or amplified fragment length polymorphism (AFLP) types. The analysis differentiated the 33 M. paratuberculosis isolates into 20 distinct MLSSR types, consistent with geographic and epidemiologic correlates and with an index of discrimination of 0.96. MLSSR analysis was also clearly able to distinguish between sheep and cattle isolates of M. paratuberculosis and easily and reproducibly differentiated strains representing the predominant MPIL genotype (genotype A18) and AFLP genotypes (genotypes Z1 and Z2) of M. paratuberculosis described previously. Taken together, the results of our studies suggest that MLSSR sequencing enables facile and reproducible high-resolution subtyping of M. paratuberculosis isolates for molecular epidemiologic and population genetic analyses.
Figures


References
-
- Bull, T. J., J. Hermon-Taylor, I. Pavlik, F. El-Zaatari, and M. Tizard. 2000. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology 146(Pt 9):2185-2197. - PubMed
-
- Bull, T. J., E. J. McMinn, K. Sidi-Boumedine, A. Skull, D. Durkin, P. Neild, G. Rhodes, R. Pickup, and J. Hermon-Taylor. 2003. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J. Clin. Microbiol. 41:2915-2923. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

