Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family
- PMID: 15071497
- PMCID: PMC394240
- DOI: 10.1038/sj.emboj.7600186
Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family
Erratum in
- EMBO J. 2005 Mar 9;24(5):1092
Abstract
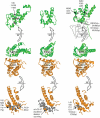
Cullin-based E3 ligases target substrates for ubiquitin-dependent degradation by the 26S proteasome. The SCF (Skp1-Cul1-F-box) and ECS (ElonginC-Cul2-SOCS box) complexes are so far the best-characterized cullin-based ligases. Their atomic structure has been solved recently, and several substrates have been described in different organisms. In addition to Cul1 and Cul2, higher eucaryotic genomes encode for three other cullins: Cul3, Cul4, and Cul5. Recent results have shed light on the molecular composition and function of Cul3-based E3 ligases. In these complexes, BTB-domain-containing proteins may bridge the cullin to the substrate in a single polypeptide, while Skp1/F-box or ElonginC/SOCS heterodimers fulfill this function in the SCF and ECS complexes. BTB-containing proteins are evolutionary conserved and involved in diverse biological processes, but their function has not previously been linked to ubiquitin-dependent degradation. In this review, we present these new findings and compare the composition of Cul3-based ligases to the well-defined SCF and ECS ligases.
Figures
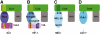

References
-
- Bomont P, Cavalier L, Blondeau F, Ben Hamida C, Belal S, Tazir M, Demir E, Topaloglu H, Korinthenberg R, Tuysuz B, Landrieu P, Hentati F, Koenig M (2000) The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet 26: 370–374 - PubMed
-
- Deffenbaugh AE, Scaglione KM, Zhang L, Moore JM, Buranda T, Sklar LA, Skowyra D (2003) Release of ubiquitin-charged Cdc34-S-Ub from the RING domain is essential for ubiquitination of the SCF(Cdc4)-bound substrate Sic1. Cell 114: 611–622 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

