Collecting, archiving and processing DNA from wildlife samples using FTA databasing paper
- PMID: 15072582
- PMCID: PMC406513
- DOI: 10.1186/1472-6785-4-4
Collecting, archiving and processing DNA from wildlife samples using FTA databasing paper
Abstract
Background: Methods involving the analysis of nucleic acids have become widespread in the fields of traditional biology and ecology, however the storage and transport of samples collected in the field to the laboratory in such a manner to allow purification of intact nucleic acids can prove problematical.
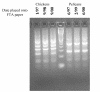
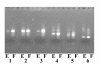
Results: FTA databasing paper is widely used in human forensic analysis for the storage of biological samples and for purification of nucleic acids. The possible uses of FTA databasing paper in the purification of DNA from samples of wildlife origin were examined, with particular reference to problems expected due to the nature of samples of wildlife origin. The processing of blood and tissue samples, the possibility of excess DNA in blood samples due to nucleated erythrocytes, and the analysis of degraded samples were all examined, as was the question of long term storage of blood samples on FTA paper. Examples of the end use of the purified DNA are given for all protocols and the rationale behind the processing procedures is also explained to allow the end user to adjust the protocols as required.
Conclusions: FTA paper is eminently suitable for collection of, and purification of nucleic acids from, biological samples from a wide range of wildlife species. This technology makes the collection and storage of such samples much simpler.
Figures
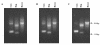
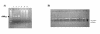



References
-
- Hsiao KM, Lin HM, Pan H, Li TC, Chen SS, Jou SB, Chiu YL, Wu MF, Lin CC, Li SY. Application of FTA® Sample Collection and DNA Purification System on the Determination of CTG Trinucleotide Repeat Size by PCR-Based Southern Blotting. Journal of Clinical Laboratory Analysis. 1999;13:188–193. doi: 10.1002/(SICI)1098-2825(1999)13:4<188::AID-JCLA8>3.0.CO;2-G. - DOI - PMC - PubMed
-
- Seah LH, Burgoyne LA. DNA data-basing on FTA® paper :Biological assault and techniques for measuring photogenic damage. In: GF Sensabaugh, PJ Lincoln, OB, editor. Progress in Forensic Genetics. Vol. 8 2000.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

