A pathway for phosphatidylcholine biosynthesis in Plasmodium falciparum involving phosphoethanolamine methylation
- PMID: 15073329
- PMCID: PMC395947
- DOI: 10.1073/pnas.0307742101
A pathway for phosphatidylcholine biosynthesis in Plasmodium falciparum involving phosphoethanolamine methylation
Abstract
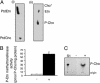
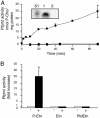
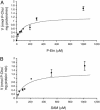
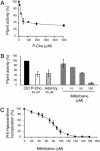
Plasmodium falciparum is the causative agent of the most severe form of human malaria. The rapid multiplication of the parasite within human erythrocytes requires an active production of new membranes. Phosphatidylcholine is the most abundant phospholipid in Plasmodium membranes, and the pathways leading to its synthesis are attractive targets for chemotherapy. In addition to its synthesis from choline, phosphatidylcholine is synthesized from serine via an unknown pathway. Serine, which is actively transported by Plasmodium from human serum and readily available in the parasite, is subsequently converted into phosphoethanolamine. Here, we describe in P. falciparum a plant-like S-adenosyl-l-methionine-dependent three-step methylation reaction that converts phosphoethanolamine into phosphocholine, a precursor for the synthesis of phosphatidylcholine. We have identified the gene, PfPMT, encoding this activity and shown that its product is an unusual phosphoethanolamine methyltransferase with no human homologs. P. falciparum phosphoethanolamine methyltransferase (Pfpmt) is a monopartite enzyme with a single catalytic domain that is responsible for the three-step methylation reaction. Interestingly, Pfpmt activity is inhibited by its product phosphocholine and by the phosphocholine analog, miltefosine. We show that miltefosine can also inhibit parasite proliferation within human erythrocytes. The importance of this enzyme in P. falciparum membrane biogenesis makes it a potential target for malaria chemotherapy.
Figures

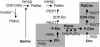
References
-
- World Health Organization (2000) WHO Tech. Rep. Ser. 892, 1-74. - PubMed
-
- Mitamura, T. & Palacpac, N. M. (2003) Microbes Infect. 5, 545-552. - PubMed
-
- Wengelnik, K., Vidal, V., Ancelin, M. L., Cathiard, A. M., Morgat, J. L., Kocken, C. H., Calas, M., Herrera, S., Thomas, A. W. & Vial, H. J. (2002) Science 295, 1311-1314. - PubMed
-
- Calas, M., Ancelin, M. L., Cordina, G., Portefaix, P., Piquet, G., Vidal-Sailhan, V. & Vial, H. (2000) J. Med. Chem. 43, 505-516. - PubMed
-
- Vial, H. J. & Ancelin, M. L. (1998) in Malaria: Parasite Biology, Pathogenesis, and Protection, ed. Shermann, I. W. (Am. Soc. Microbiol. Press, Washington, DC), pp. 159-175.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

