Tfs1p, a member of the PEBP family, inhibits the Ira2p but not the Ira1p Ras GTPase-activating protein in Saccharomyces cerevisiae
- PMID: 15075275
- PMCID: PMC387632
- DOI: 10.1128/EC.3.2.459-470.2004
Tfs1p, a member of the PEBP family, inhibits the Ira2p but not the Ira1p Ras GTPase-activating protein in Saccharomyces cerevisiae
Abstract
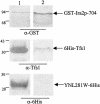
Ras proteins are guanine nucleotide-binding proteins that are highly conserved among eukaryotes. They are involved in signal transduction pathways and are tightly regulated by two sets of antagonistic proteins: GTPase-activating proteins (GAPs) inhibit Ras proteins, whereas guanine exchange factors activate them. In this work, we describe Tfs1p, the first physiological inhibitor of a Ras GAP, Ira2p, in Saccharomyces cerevisiae. TFS1 is a multicopy suppressor of the cdc25-1 mutation in yeast and corresponds to the so-called Ic CPY cytoplasmic inhibitor. Moreover, Tfs1p belongs to the phosphatidylethanolamine-binding protein (PEBP) family, one member of which is RKIP, a kinase and serine protease inhibitor and a metastasis inhibitor in prostate cancer. In this work, the results of (i) a two-hybrid screen of a yeast genomic library, (ii) glutathione S-transferase pulldown experiments, (iii) multicopy suppressor tests of cdc25-1 mutants, and (iv) stress resistance tests to evaluate the activation level of Ras demonstrate that Tfs1p interacts with and inhibits Ira2p. We further show that the conserved ligand-binding pocket of Tfs1-the hallmark of the PEBP family-is important for its inhibitory activity.
Figures




References
-
- Ahmadian, M. R., P. Stege, K. Scheffzek, and A. Wittinghofer. 1997. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 4:686-689. - PubMed
-
- Banfield, M. J., J. J. Barker, A. C. Perry, and R. L. Brady. 1998. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure 6:1245-1254. - PubMed
-
- Banfield, M. J., and R. L. Brady. 2000. The structure of Antirrhinum centroradialis protein (CEN) suggests a role as a kinase regulator. J. Mol. Biol. 297:1159-1170. - PubMed
-
- Boguski, M. S., and F. McCormick. 1993. Proteins regulating Ras and its relatives. Nature 366:643-654. - PubMed
-
- Bollag, G., and F. McCormick. 1991. Differential regulation of rasGAP and neurofibromatosis gene product activities. Nature 351:576-579. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

