Cryptosporidium parvum mitochondrial-type HSP70 targets homologous and heterologous mitochondria
- PMID: 15075277
- PMCID: PMC387664
- DOI: 10.1128/EC.3.2.483-494.2004
Cryptosporidium parvum mitochondrial-type HSP70 targets homologous and heterologous mitochondria
Abstract
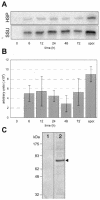
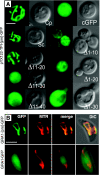
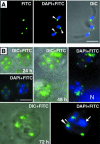
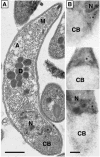
A mitochondrial HSP70 gene (Cp-mtHSP70) is described for the apicomplexan Cryptosporidium parvum, an agent of diarrhea in humans and animals. Mitochondrial HSP70 is known to have been acquired from the proto-mitochondrial endosymbiont. The amino acid sequence of Cp-mtHSP70 shares common domains with mitochondrial and proteobacterial homologues, including 34 amino acids of an NH2-terminal mitochondrion-like targeting presequence. Phylogenetic reconstruction places Cp-mtHSP70 within the mitochondrial clade of HSP70 homologues. Using reverse transcription-PCR, Cp-mtHSP70 mRNA was observed in C. parvum intracellular stages cultured in HCT-8 cells. Polyclonal antibodies to Cp-mtHSP70 recognize a approximately 70-kDa protein in Western blot analysis of sporozoite extracts. Both fluorescein- and immunogold-labeled anti-Cp-mtHSP70 localize to a single mitochondrial compartment in close apposition to the nucleus. Furthermore, the NH2-terminal presequence of Cp-mtHSP70 can correctly target green fluorescent protein to the single mitochondrion of the apicomplexan Toxoplasma gondii and the mitochondrial network of the yeast Saccharomyces cerevisiae. When this presequence was truncated, the predicted amphiphilic alpha-helix was shown to be essential for import into the yeast mitochondrion. These data further support the presence of a secondarily reduced relict mitochondrion in C. parvum.
Figures



References
-
- Abe, Y., T. Shodai, T. Muto, K. Mihara, H. Torii, S. Nishikawa, T. Endo, and D. Kohda. 2000. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100:551-560. - PubMed
-
- Abrahamsen, M. S., and A. A. Schroeder. 1999. Characterization of intracellular Cryptosporidium parvum gene expression. Mol. Biochem. Parasitol. 104:141-146. - PubMed
-
- Alvarez-Pellitero, P., and A. Sitja-Bobadilla. 2002. Cryptosporidium molnari n. sp. (Apicomplexa: Cryptosporidiidae) infecting two marine fish species, Sparus aurata L. and Dicentrarchus labrax L. Int. J. Parasitol. 32:1007-1021. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

