Critical nucleation size in the folding of small apparently two-state proteins
- PMID: 15075405
- PMCID: PMC2286761
- DOI: 10.1110/ps.03587604
Critical nucleation size in the folding of small apparently two-state proteins
Abstract
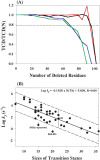
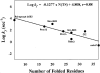
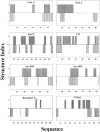
For apparently two-state proteins, we found that the size (number of folded residues) of a transition state is mostly encoded by the topology, defined by total contact distance (TCD) of the native state, and correlates with its folding rate. This is demonstrated by using a simple procedure to reduce the native structures of the 41 two-state proteins with native TCD as a constraint, and is further supported by analyzing the results of eight proteins from protein engineering studies. These results support the hypothesis that the major rate-limiting process in the folding of small apparently two-state proteins is the search for a critical number of residues with the topology close to that of the native state.
Figures

References
-
- Alm, E., Morozov, A.V., Kortemme, T., and Baker, D. 2002. Simple physical models connect theory and experiment in protein folding kinetics. J. Mol.Biol. 322 463–476. - PubMed
-
- Baldwin, R.L. and Rose, G.D. 1999. Is protein folding hierarchic? I. Local structure and peptide folding. Trends Biochem. Sci. 24 26–33. - PubMed
-
- Chiti, F., Taddei, N., White, P.M., Bucciantini, M., Magherini, F., Stefani, M., and Dobson, C.M. 1999. Mutational analysis of acylphosphatase suggests the importance of topology and contact order in protein folding. Nat. Struct. Biol. 6 1005–1009. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

