Krüppel-like factor 5 mediates the transforming activity of oncogenic H-Ras
- PMID: 15077182
- PMCID: PMC1351030
- DOI: 10.1038/sj.onc.1207397
Krüppel-like factor 5 mediates the transforming activity of oncogenic H-Ras
Abstract
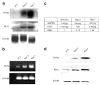
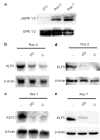
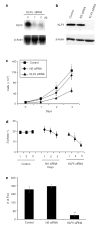
Previous studies indicate that Krüppel-like factor 5 (KLF5), also known as intestinal-enriched Krüppel-like factor (IKLF), is a positive regulator of cell proliferation and gives rise to a transformed phenotype when overexpressed. Here we demonstrate that levels of KLF5 transcript and protein are significantly elevated in oncogenic H-Ras-transformed NIH3T3 cells. These cells display an accelerated rate of proliferation in both serum-containing and serum-deprived media and form anchorage-independent colonies in soft agar assays. H-Ras-transformed cells also contain elevated mitogen-activated protein kinase (MAPK) activity. When treated with inhibitors of MEK (MAPK kinase), H-Ras-transformed cells lose their growth advantage and no longer form colonies. Significantly, levels of KLF5 transcript and protein are substantially reduced in H-Ras-transformed cells treated with MEK inhibitors. Moreover, inhibition of KLF5 expression in H-Ras-transformed cells with KLF5-specific small interfering RNA (siRNA) leads to a decreased rate of proliferation and a significant reduction in colony formation. H-Ras-transformed cells also contain elevated levels of Egr1 that are diminished by MEK inhibitors. Inhibition of Egr1 by siRNA results in a reduced level of KLF5, indicating that Egr1 mediates the inductive action of MAPK on KLF5. Lastly, KLF5 activates expression of cyclin D1. These findings indicate that the increased expression of KLF5 in H-Ras-transformed cells is secondary to increased MAPK activity from H-Ras overexpression and that the elevated level of KLF5 is in part responsible for the proproliferative and transforming activities of oncogenic H-Ras.
Copyright 2004 Nature Publishing Group
Figures





References
-
- Barbacid M. Annu Rev Biochem. 1987;56:779–827. - PubMed
-
- Black AR, Black JD, Azizkhan-Clifford J. J Cell Physiol. 2001;188:143–160. - PubMed
-
- Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, Vogelstein B. Nature. 1987;327:293–297. - PubMed
-
- Chen C, Bhalala HV, Qiao H, Dong JT. Oncogene. 2002;21:6567–6572. - PubMed
-
- Chen C, Bhalala HV, Vessella RL, Dong JT. Prostate. 2003;55:81–88. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

