Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo
- PMID: 15078818
- PMCID: PMC395847
- DOI: 10.1101/gad.1184604
Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo
Abstract
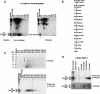
Posttranslational histone modifications are important for the regulation of many biological phenomena. Here, we show the purification and characterization of nucleosomal histone kinase-1 (NHK-1). NHK-1 has a high affinity for chromatin and phosphorylates a novel site, Thr 119, at the C terminus of H2A. Notably, NHK-1 specifically phosphorylates nucleosomal H2A, but not free H2A in solution. In Drosophila embryos, phosphorylated H2A Thr 119 is found in chromatin, but not in the soluble core histone pool. Immunostaining of NHK-1 revealed that it goes to chromatin during mitosis and is excluded from chromatin during S phase. Consistent with the shuttling of NHK-1 between chromatin and cytoplasm, H2A Thr 119 is phosphorylated during mitosis but not in S phase. These studies reveal that NHK-1-catalyzed phosphorylation of a conserved serine/threonine residue in H2A is a new component of the histone code that might be related to cell cycle progression.
Figures
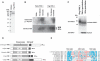
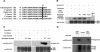
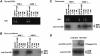


References
-
- Anest, V., Hanson, J.L., Cogswell, P.C., Steinbrecher, K.A., Strahl, B.D., and Baldwin, A.S. 2003. A nucleosomal function for IκB kinase-α in NF-κB-dependent gene expression. Nature 423: 659–663. - PubMed
-
- Ballal, N.R., Kang, Y.J., Olson, M.O., and Busch, H. 1975. Changes in nucleolar proteins and their phosphorylation patterns during liver regeneration. J. Biol. Chem. 250: 5921–5925. - PubMed
-
- Bannister, A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. - PubMed
-
- Carrozza, M.J., Utley, R.T., Workman, J.L., and Cote, J. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19: 321–329. - PubMed
-
- Cheung, P., Tanner, K.G., Cheung, W.L., Sassone-Corsi, P., Denu, J.M., and Allis, C.D. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5: 905–915. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
