Comparison of human chromosome 21 conserved nongenic sequences (CNGs) with the mouse and dog genomes shows that their selective constraint is independent of their genic environment
- PMID: 15078857
- PMCID: PMC479112
- DOI: 10.1101/gr.1934904
Comparison of human chromosome 21 conserved nongenic sequences (CNGs) with the mouse and dog genomes shows that their selective constraint is independent of their genic environment
Abstract
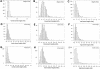
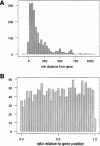
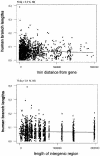
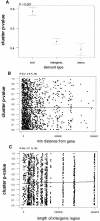
The analysis of conservation between the human and mouse genomes resulted in the identification of a large number of conserved nongenic sequences (CNGs). The functional significance of this nongenic conservation remains unknown, however. The availability of the sequence of a third mammalian genome, the dog, allows for a large-scale analysis of evolutionary attributes of CNGs in mammals. We have aligned 1638 previously identified CNGs and 976 conserved exons (CODs) from human chromosome 21 (Hsa21) with their orthologous sequences in mouse and dog. Attributes of selective constraint, such as sequence conservation, clustering, and direction of substitutions were compared between CNGs and CODs, showing a clear distinction between the two classes. We subsequently performed a chromosome-wide analysis of CNGs by correlating selective constraint metrics with their position on the chromosome and relative to their distance from genes. We found that CNGs appear to be randomly arranged in intergenic regions, with no bias to be closer or farther from genes. Moreover, conservation and clustering of substitutions of CNGs appear to be completely independent of their distance from genes. These results suggest that the majority of CNGs are not typical of previously described regulatory elements in terms of their location. We propose models for a global role of CNGs in genome function and regulation, through long-distance cis or trans chromosomal interactions.
Figures

References
-
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391-1394. - PubMed
-
- Dermitzakis, E.T., Reymond, A., Lyle, R., Scamuffa, N., Ucla, C., Deutsch, S., Stevenson, B.J., Flegel, V., Bucher, P., Jongeneel, C.V., et al. 2002. Numerous potentially functional but nongenic conserved sequences on human chromosome 21. Nature 420: 578-582. - PubMed
-
- Dermitzakis, E.T., Reymond, A., Scamuffa, N., Ucla, C., Kirkness, E., Rossier, C., and Antonarakis, S.E. 2003. Evolutionary discrimination of mammalian conserved nongenic sequences (CNGs). Science 302: 1033-1035. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
