Regulation of spalt expression in the Drosophila wing blade in response to the Decapentaplegic signaling pathway
- PMID: 15079076
- PMCID: PMC395916
- DOI: 10.1073/pnas.0401590101
Regulation of spalt expression in the Drosophila wing blade in response to the Decapentaplegic signaling pathway
Abstract
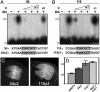
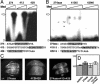
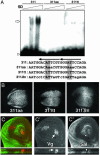
Pattern formation depends on the acquisition of precise cellular identities due to the differential expression of transcription factors. Enhancers within regulatory regions integrate the positive and negative regulatory signals directing gene transcription. Here, we analyze the enhancer that drives expression of the Drosophila gene spalt in the wing blade. This enhancer integrates positive signals, mediated by the Decapentaplegic signaling effector protein Medea, with the repressor activity of Brinker. The enhancer functions in the absence of binding sites for the wing-specific factor Scalloped. The molecular analysis of this enhancer indicates that there are additional factors yet unknown involved in the activation of spalt in the wing blade and that the mechanism of repression by Brinker does not rely on competition with Mad-Medea overlapping sites. The comparisons with other enhancers that respond to Decapentaplegic suggest that there are different possibilities to integrate the positive and negative inputs triggered by this signaling pathway.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

