Relevance of connexin deafness (DFNB1) to human evolution
- PMID: 15079193
- PMCID: PMC1182073
- DOI: 10.1086/420979
Relevance of connexin deafness (DFNB1) to human evolution
Abstract
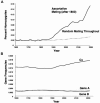
The connexins are the subunits of a family of proteins that form gap junctions, allowing ions and small molecules to move between adjacent cells. At least four connexins are expressed in the ear, and, although there are known mutations at >100 loci that can cause deafness, those involving DFNB1, in the interval 13q11-q12 containing the GJB2 and GJB6 genes coding for connexins 26 and 30, are the most frequent cause of recessive deafness in many populations. We have suggested that the combined effects of relaxed selection and linguistic homogamy can explain the high frequency of connexin deafness and may have doubled its incidence in this country during the past 200 years. In this report, we show by computer simulation that assortative mating, in fact, can accelerate dramatically the genetic response to relaxed selection. Along with the effects of gene drift and consanguinity, assortative mating also may have played a key role in the joint evolution and accelerated fixation of genes for speech after they first appeared in Homo sapiens 100,000-150,000 years ago.
Figures
References
Electronic-Database Information
-
- Hereditary Hearing Loss, http://www.uia.ac.be/dnalab/hhh/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM=Limits
References
-
- Bender RF (1970) The conquest of deafness. Case Western Reserve, Cleveland, pp 1–243
-
- Clark AG, Glanowski S, Neilsen R, Thomas PD, Kejarwal A, Todd MA, Tannenbaum DM, Civello D, Lu F, Murphy B, Ferriera S, Wang G, Zheng X, White TJ, Sninsky JJ, Adams MD, Cargill M (2003) Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science 302:1960–1963 10.1126/science.1088821 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

