Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor
- PMID: 15082532
- PMCID: PMC387422
- DOI: 10.1101/gad.1200704
Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor
Abstract
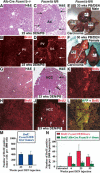
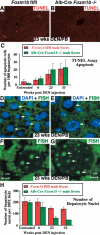
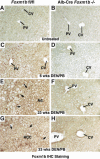
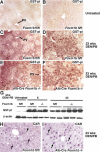
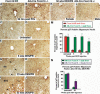
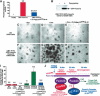
Hepatocellular carcinoma (HCC) is a leading cause of cancer-related deaths worldwide. Here, we provide evidence that the Forkhead Box (Fox) m1b (Foxm1b or Foxm1) transcription factor is essential for the development of HCC. Conditionally deleted Foxm1b mouse hepatocytes fail to proliferate and are highly resistant to developing HCC in response to a Diethylnitrosamine (DEN)/Phenobarbital (PB) liver tumor-induction protocol. The mechanism of resistance to HCC development is associated with nuclear accumulation of the cell cycle inhibitor p27(Kip1) protein and reduced expression of the Cdk1-activator Cdc25B phosphatase. We showed that the Foxm1b transcription factor is a novel inhibitory target of the p19(ARF) tumor suppressor. Furthermore, we demonstrated that conditional overexpression of Foxm1b protein in osteosarcoma U2OS cells greatly enhances anchorage-independent growth of cell colonies on soft agar. A p19(ARF) 26-44 peptide containing nine D-Arg to enhance cellular uptake of the peptide was sufficient to significantly reduce both Foxm1b transcriptional activity and Foxm1b-induced growth of U2OS cell colonies on soft agar. These results suggest that this (D-Arg)(9)-p19(ARF) 26-44 peptide is a potential therapeutic inhibitor of Foxm1b function during cellular transformation. Our studies demonstrate that the Foxm1b transcription factor is required for proliferative expansion during tumor progression and constitutes a potential new target for therapy of human HCC tumors.
Figures



References
-
- Barr F.G. 2001. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene 20: 5736-5746. - PubMed
-
- Barr F.G., Nauta, L.E., Davis, R.J., Schafer, B.W., Nycum, L.M., and Biegel, J.A. 1996. In vivo amplification of the PAX3–FKHR and PAX7–FKHR fusion genes in alveolar rhabdomyosarcoma. Hum. Mol. Genet. 5: 15-21. - PubMed
-
- Becker-Hapak M., McAllister, S.S., and Dowdy, S.F. 2001. TAT-mediated protein transduction into mammalian cells. Methods 24: 247-256. - PubMed
-
- Block T.M., Mehta, A.S., Fimmel, C.J., and Jordan, R. 2003. Molecular viral oncology of hepatocellular carcinoma. Oncogene 22: 5093-5107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
