Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease
- PMID: 15082587
- PMCID: PMC1774050
- DOI: 10.1136/gut.2003.025403
Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease
Abstract
Background and aims: The intestinal bacterial microflora plays an important role in the aetiology of inflammatory bowel disease (IBD). As most of the colonic bacteria cannot be identified by culture techniques, genomic technology can be used for analysis of the composition of the microflora.
Patients and methods: The mucosa associated colonic microflora of 57 patients with active inflammatory bowel disease and 46 controls was investigated using 16S rDNA based single strand conformation polymorphism (SSCP) fingerprint, cloning experiments, and real time polymerase chain reaction (PCR).
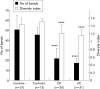
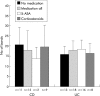
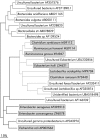
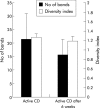
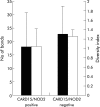
Results: Full length sequencing of 1019 clones from 16S rDNA libraries (n = 3) revealed an overall bacterial diversity of 83 non-redundant sequences-among them, only 49 known bacterial species. Molecular epidemiology of the composition of the colonic microflora was investigated by SSCP. Diversity of the microflora in Crohn's disease was reduced to 50% compared with controls (21.7 v 50.4; p<0.0001) and to 30% in ulcerative colitis (17.2 v 50.4; p<0.0001). The reduction in diversity in inflammatory bowel disease was due to loss of normal anaerobic bacteria such as Bacteroides species, Eubacterium species, and Lactobacillus species, as revealed by direct sequencing of variable bands and confirmed by real time PCR. Bacterial diversity in the Crohn's group showed no association with CARD15/NOD2 status.
Conclusions: Mucosal inflammation in inflammatory bowel disease is associated with loss of normal anaerobic bacteria. This effect is independent of NOD2/CARD15 status of patients.
Figures


References
-
- Hooper LV, Wong MH, Thelin A, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science 2001;291:881–4. - PubMed
-
- Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science 2001;292:1115–18. - PubMed
-
- Madsen KL, Doyle JS, Jewell LD, et al. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology 1999;116:1107–14. - PubMed
-
- Hampe J, Cuthbert A, Croucher PJ, et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet 2001;357:1925–8. - PubMed
-
- Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001;411:599–603. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
