The Apc5 subunit of the anaphase-promoting complex/cyclosome interacts with poly(A) binding protein and represses internal ribosome entry site-mediated translation
- PMID: 15082755
- PMCID: PMC387753
- DOI: 10.1128/MCB.24.9.3577-3587.2004
The Apc5 subunit of the anaphase-promoting complex/cyclosome interacts with poly(A) binding protein and represses internal ribosome entry site-mediated translation
Abstract
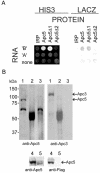
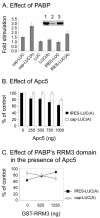
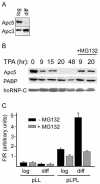
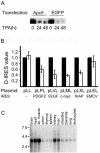
The anaphase-promoting complex/cyclosome (APC/C) is a multisubunit ubiquitin ligase that mediates the proteolysis of cell cycle proteins in mitosis and G(1). We used a yeast three-hybrid screen to identify proteins that interact with the internal ribosome entry site (IRES) of platelet-derived growth factor 2 mRNA. Surprisingly, this screen identified Apc5, although it does not harbor a classical RNA binding domain. We found that Apc5 binds the poly(A) binding protein (PABP), which directly binds the IRES element. PABP was found to enhance IRES-mediated translation, whereas Apc5 overexpression counteracted this effect. In addition to its association with the APC/C complex, Apc5 binds much heavier complexes and cosediments with the ribosomal fraction. In contrast to Apc3, which is associated only with the APC/C and remains intact during differentiation, Apc5 is degraded upon megakaryocytic differentiation in correlation with IRES activation. Expression of Apc5 in differentiated cells abolished IRES activation. This is the first report implying an additional role for an APC/C subunit, apart from its being part of the APC/C complex.
Figures




References
-
- Bentley, A. M., B. C. Williams, M. L. Goldberg, and A. J. Andres. 2002. Phenotypic characterization of Drosophila ida mutants: defining the role of APC5 in cell cycle progression. J. Cell Sci. 115:949-961. - PubMed
-
- Bernstein, J., O. Sella, S. Y. Le, and O. Elroy-Stein. 1997. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES). J. Biol. Chem. 272:9356-9362. - PubMed
-
- Cornelis, S., Y. Bruynooghe, G. Denecker, S. Van Huffel, S. Tinton, and R. Beyaert. 2000. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell 5:597-605. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
