Protein kinase C-mediated phosphorylation of the leukemia-associated HOXA9 protein impairs its DNA binding ability and induces myeloid differentiation
- PMID: 15082777
- PMCID: PMC387750
- DOI: 10.1128/MCB.24.9.3827-3837.2004
Protein kinase C-mediated phosphorylation of the leukemia-associated HOXA9 protein impairs its DNA binding ability and induces myeloid differentiation
Abstract
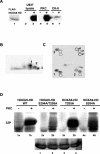
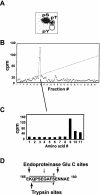
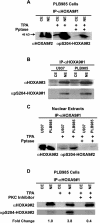
HOXA9 expression is a common feature of acute myeloid leukemia, and high-level expression is correlated with poor prognosis. Moreover, HOXA9 overexpression immortalizes murine marrow progenitors that are arrested at a promyelocytic stage of differentiation when cultured and causes leukemia in recipient mice following transplantation of HOXA9 expressing bone marrow. The molecular mechanisms underlying the physiologic functions and transforming properties of HOXA9 are poorly understood. This study demonstrates that HOXA9 is phosphorylated by protein kinase C (PKC) and casein kinase II and that PKC mediates phosphorylation of purified HOXA9 on S204 as well as on T205, within a highly conserved consensus sequence, in the N-terminal region of the homeodomain. S204 in the endogenous HOXA9 protein was phosphorylated in PLB985 myeloid cells, as well as in HOXA9-immortalized murine marrow cells. This phosphorylation was enhanced by phorbol ester, a known inducer of PKC, and was inhibited by a specific PKC inhibitor. PKC-mediated phosphorylation of S204 decreased HOXA9 DNA binding affinity in vitro and the ability of the endogenous HOXA9 to form cooperative DNA binding complexes with PBX. PKC inhibition significantly reduced the phorbol-ester induced differentiation of the PLB985 hematopoietic cell line as well as HOXA9-immortalized murine bone marrow cells. These data suggest that phorbol ester-induced myeloid differentiation is in part due to PKC-mediated phosphorylation of HOXA9, which decreases the DNA binding of the homeoprotein.
Figures


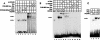

Similar articles
-
Nup98-HoxA9 immortalizes myeloid progenitors, enforces expression of Hoxa9, Hoxa7 and Meis1, and alters cytokine-specific responses in a manner similar to that induced by retroviral co-expression of Hoxa9 and Meis1.Oncogene. 2002 Jun 20;21(27):4247-56. doi: 10.1038/sj.onc.1205516. Oncogene. 2002. PMID: 12082612
-
Characterization of IkappaB kinases. IkappaB-alpha is not phosphorylated by Raf-1 or protein kinase C isozymes, but is a casein kinase II substrate.J Biol Chem. 1996 Jun 7;271(23):13868-74. doi: 10.1074/jbc.271.23.13868. J Biol Chem. 1996. PMID: 8662925
-
Phosphorylation of murine homeodomain protein Dlx3 by protein kinase C.FEBS Lett. 2001 May 4;496(1):60-5. doi: 10.1016/s0014-5793(01)02398-5. FEBS Lett. 2001. PMID: 11343707 Free PMC article.
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
-
The potential for isoenzyme-selective modulation of protein kinase C.FASEB J. 1997 Jul;11(8):649-69. doi: 10.1096/fasebj.11.8.9240967. FASEB J. 1997. PMID: 9240967 Review.
Cited by
-
Mechanisms of Specificity for Hox Factor Activity.J Dev Biol. 2016 Jun;4(2):16. doi: 10.3390/jdb4020016. Epub 2016 May 9. J Dev Biol. 2016. PMID: 27583210 Free PMC article.
-
Phosphorylation of the leukemic oncoprotein EVI1 on serine 196 modulates DNA binding, transcriptional repression and transforming ability.PLoS One. 2013 Jun 12;8(6):e66510. doi: 10.1371/journal.pone.0066510. Print 2013. PLoS One. 2013. PMID: 23776681 Free PMC article.
-
Polycomb repressor complex 2 regulates HOXA9 and HOXA10, activating ID2 in NK/T-cell lines.Mol Cancer. 2010 Jun 17;9:151. doi: 10.1186/1476-4598-9-151. Mol Cancer. 2010. PMID: 20565746 Free PMC article.
-
HOXA9 participates in the transcriptional activation of E-selectin in endothelial cells.Mol Cell Biol. 2007 Jun;27(12):4207-16. doi: 10.1128/MCB.00052-07. Epub 2007 Apr 23. Mol Cell Biol. 2007. PMID: 17452460 Free PMC article.
-
Direct and Indirect Targeting of HOXA9 Transcription Factor in Acute Myeloid Leukemia.Cancers (Basel). 2019 Jun 17;11(6):837. doi: 10.3390/cancers11060837. Cancers (Basel). 2019. PMID: 31213012 Free PMC article. Review.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology, p. 12.1.1-12.1.8. John Wiley & Sons, Hoboken, N.J.
-
- Borrow, J., A. M. Shearman, V. P. J. Stanton, R. Becher, T. Collins, A. J. Williams, I. Dube, F. Katz, Y. L. Kwong, C. Morris, K. Ohyashiki, K. Toyama, J. Rowley, and D. E. Housman. 1996. The t(7;11)(p15;p15) translocation in acute myeloid leukemia fuses the genes for nucleoporin NU98 and class I homeoprotein HOXA9. Nat. Genet. 12:159-167. - PubMed
-
- Bourbon, H. M., E. Martin-Blanco, D. Rosen, and T. B. Kornberg. 1995. Phosphorylation of the Drosophila engrailed protein at a site outside its homeodomain enhances DNA binding. J. Biol. Chem. 270:11130-11139. - PubMed
-
- Boyle, W. J., P. van der Geer, and T. Hunter. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201:110-143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
