The NRIF3 family of transcriptional coregulators induces rapid and profound apoptosis in breast cancer cells
- PMID: 15082778
- PMCID: PMC387764
- DOI: 10.1128/MCB.24.9.3838-3848.2004
The NRIF3 family of transcriptional coregulators induces rapid and profound apoptosis in breast cancer cells
Abstract
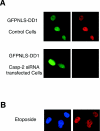
Many anticancer drugs kill cancer cells by inducing apoptosis. Despite the progress in understanding apoptosis, how to harness the cellular death machinery to selectively deliver tumor-specific cytotoxicity (while minimizing damage to other cells) remains an important challenge. We report here that expression of the NRIF3 family of transcriptional coregulators in a variety of breast cancer cell lines induces rapid and profound apoptosis (nearly 100% cell death within 24 h). A novel death domain (DD1) was mapped to a short 30-amino-acid region common to all members of the NRIF3 family. Mechanistic studies showed that DD1-induced apoptosis occurs through a novel caspase 2-mediated pathway that involves mitochondrial membrane permeabilization but does not require other caspases. Interestingly, the cytotoxicity of NRIF3 and DD1 appears to be cell type specific, as they selectively kill breast cancer or related cells but not other examined cells of different origins. Our study demonstrates the feasibility of selectively inducing cytotoxicity in a specific cancer and suggests that breast cancer cells contain a novel "death switch" that can be specifically triggered by NRIF3 or DD1. Strategies utilizing NRIF3 and/or DD1 and/or targeting this death switch may lead to the development of novel and more selective therapeutics against breast cancer.
Figures















References
-
- Adam, S. A., and L. Gerace. 1991. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell 66:837-847. - PubMed
-
- Afonja, A., B. M. Raaka, A. Huang, S. Das, X. Zhao, E. Helmer, D. Juste, and H. H. Samuels. 2002. RAR agonists stimulate SOX9 gene expression in breast cancer cell lines: evidence for a role in retinoid-mediated growth inhibition. Oncogene 21:7850-7860. - PubMed
-
- Antonsson, B. 2001. Bax and other pro-apoptotic Bcl-2 family “killer-proteins” and their victim the mitochondrion. Cell Tissue Res. 306:347-361. - PubMed
-
- Baliga, B. C., P. A. Colussi, S. H. Read, M. M. Dias, D. A. Jans, and S. Kumar. 2003. Role of prodomain in importin-mediated nuclear localization and activation of caspase-2. J. Biol. Chem. 278:4899-4905. - PubMed
-
- Benson, J. R., and V. Pitsinis. 2003. Update on clinical role of tamoxifen. Curr. Opin. Obstet. Gynecol. 15:13-23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
