Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells
- PMID: 15084673
- PMCID: PMC390380
- DOI: 10.1093/nar/gnh063
Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells
Abstract
Real-time visualization of specific endogenous mRNA expression in vivo has the potential to revolutionize medical diagnosis, drug discovery, developmental and molecular biology. However, conventional liposome- or dendrimer-based cellular delivery of molecular probes is inefficient, slow, and often detrimental to the probes. Here we demonstrate the rapid and sensitive detection of RNA in living cells using peptide-linked molecular beacons that possess self-delivery, targeting and reporting functions. We conjugated the TAT peptide to molecular beacons using three different linkages and demonstrated that, at relatively low concentrations, these molecular beacon constructs were internalized into living cells within 30 min with nearly 100% efficiency. Further, peptide-based delivery did not interfere with either specific targeting by or hybridization-induced fluorescence of the probes. We could therefore detect human GAPDH and survivin mRNAs in living cells fluorescently, revealing intriguing intracellular localization patterns of mRNA. We clearly demonstrated that cellular delivery of molecular beacons using the peptide-based approach has far better performance compared with conventional transfection methods. The peptide-linked molecular beacons approach promises to open new and exciting opportunities in sensitive gene detection and quantification in vivo.
Figures

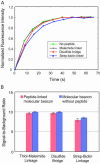



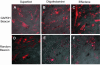
References
-
- Karge W.H. 3rd, Schaefer,E.J. and Ordovas,J.M. (1998) Quantification of mRNA by polymerase chain reaction (PCR) using an internal standard and a nonradioactive detection method. Methods Mol. Biol., 110, 43–61. - PubMed
-
- Jung R., Soondrum,K. and Neumaier,M. (2000) Quantitative PCR. Clin. Chem. Lab. Med., 38, 833–836. - PubMed
-
- Brown P.O. and Botstein,D. (1999) Exploring the new world of the genome with DNA microarrays. Nature Genet., 21, 33–37. - PubMed
-
- DeRisi J.L. and Iyer,V.R. (1999) Genomics and array technology. Curr. Opin. Oncol., 11, 76–79. - PubMed
-
- Femino A.M., Fay,F.S., Fogarty,K. and Singer,R.H. (1998) Visualization of single RNA transcripts in situ. Science, 280, 585–590. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

