The GATA family of transcription factors in Arabidopsis and rice
- PMID: 15084732
- PMCID: PMC419845
- DOI: 10.1104/pp.103.037788
The GATA family of transcription factors in Arabidopsis and rice
Abstract
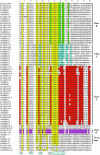
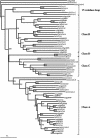
GATA transcription factors are a group of DNA binding proteins broadly distributed in eukaryotes. The GATA factors DNA binding domain is a class IV zinc finger motif in the form CX(2)CX(17-20)CX(2)C followed by a basic region. In plants, GATA DNA motifs have been implicated in light-dependent and nitrate-dependent control of transcription. Herein, we show that the Arabidopsis and the rice (Oryza sativa) genomes present 29 and 28 loci, respectively, that encode for putative GATA factors. A phylogenetic analysis of the 57 GATA factors encoding genes, as well as the study of their intron-exon structure, indicates the existence of seven subfamilies of GATA genes. Some of these subfamilies are represented in both species but others are exclusive for one of them. In addition to the GATA zinc finger motif, polypeptides of the different subfamilies are characterized by the presence of additional domains such as an acidic domain, a CCT (CONSTANS, CO-like, and TOC1) domain, or a transposase-like domain also found in FAR1 and FHY3. Subfamily VI comprises genes that encode putative bi-zinc finger polypeptides, also found in metazoan and fungi, and a tri-zinc finger protein which has not been previously reported in eukaryotes. The phylogeny of the GATA zinc finger motif, excluding flanking regions, evidenced the existence of four classes of GATA zinc fingers, three of them containing 18 residues in the zinc finger loop and one containing a 20-residue loop. Our results support multiple models of evolution of the GATA gene family in plants including gene duplication and exon shuffling.
Figures





References
-
- Argüello-Astorga G, Herrera-Estrella L (1998) Evolution of light-regulated plant promoters. Annu Rev Plant Physiol Plant Mol Biol 49: 525–555 - PubMed
-
- Borello U, Ceccarelli E, Giuliano G (1993) Constitutive, light-responsive and circadian clock-responsive factors compete for the different l box elements in plant light-regulated promoters. Plant J 4: 611–619 - PubMed
-
- Caddick MX, Arst HN, Jr. (1990) Nitrogen regulation in Aspergillus: are two fingers better than one? Gene 95: 123–127 - PubMed
-
- Castresana C, Staneloni RJ, Malik VS, Cashmore AR (1987) Molecular characterization of two clusters of genes encoding the Type I CAB polypeptides of PSII in Nicotiana plumbaginifolia. Plant Mol Biol 10: 117–126 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

