Human centromere repositioning "in progress"
- PMID: 15084747
- PMCID: PMC404081
- DOI: 10.1073/pnas.0308637101
Human centromere repositioning "in progress"
Abstract
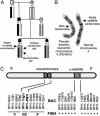
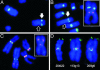
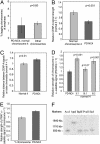
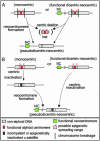
Centromere repositioning provides a potentially powerful evolutionary force for reproductive isolation and speciation, but the underlying mechanisms remain ill-defined. An attractive model is through the simultaneous inactivation of a normal centromere and the formation of a new centromere at a hitherto noncentromeric chromosomal location with minimal detrimental effect. We report a two-generation family in which the centromeric activity of one chromosome 4 has been relocated to a euchromatic site at 4q21.3 through the epigenetic formation of a neocentromere in otherwise cytogenetically normal and mitotically stable karyotypes. Strong epigenetic inactivation of the original centromere is suggested by retention of 1.3 megabases of centromeric alpha-satellite DNA, absence of detectable molecular alteration in chromosome 4-centromereproximal p- and q-arm sequences, and failure of the inactive centromere to be reactivated through extensive culturing or treatment with histone deacetylase inhibitor trichostatin A. The neocentromere binds functionally essential centromere proteins (CENP-A, CENP-C, CENP-E, CENP-I, BUB1, and HP1), although a moderate reduction in CENP-A binding and sister-chromatid cohesion compared with the typical centromeres suggests possible underlying structural/functional differences. The stable mitotic and meiotic transmissibility of this pseudodicentric-neocentric chromosome in healthy individuals and the ability of the neocentric activity to form in a euchromatic site in preference to a preexisting alphoid domain provide direct evidence for an inherent mechanism of human centromere repositioning and karyotype evolution "in progress." We discuss the wider implication of such a mechanism for meiotic drive and the evolution of primate and other species.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

