Regulation of ultraviolet light-induced gene expression by gene size
- PMID: 15087501
- PMCID: PMC404088
- DOI: 10.1073/pnas.0308181101
Regulation of ultraviolet light-induced gene expression by gene size
Abstract
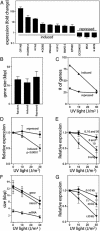
UV light induces the expression of a wide variety of genes. At present, it is unclear how cells sense the extent of DNA damage and alter the expression of UV-induced genes appropriately. UV light induces DNA damage that blocks transcription, and the probability that a gene sustains transcription-blocking DNA damage is proportional to locus size and dose of UV light. Using colon carcinoma cells that express a temperature-sensitive variant of p53 and undergo p53-dependent apoptosis after UV irradiation, we found that the number of p53-induced genes identified by oligonucleotide microarray analysis decreased in a UV dose-dependent manner. This was associated with a statistically significant shift in the spectrum of p53-induced genes toward compact genes with fewer and smaller introns. Genes encoding proapoptotic proteins involved in the initiation of the mitochondrial apoptotic cascade were prominent among the compact p53 target genes, whereas genes encoding negative regulators of p53 and the mitochondrial apoptotic pathway were significantly larger. We propose that the shift in spectrum of UV-responsive gene expression caused by passive effects of UV lesions on transcription acts as a molecular dosimeter, ensuring the elimination of cells sustaining irreparable transcription-blocking DNA damage.
Figures

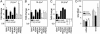
References
-
- Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1351. - PubMed
-
- Tornaletti, S. & Hanawalt, P. (1999) Biochimie 81, 139-146. - PubMed
-
- Mayne, L. & Lehmann, A. (1982) Cancer Res. 42, 1473-1478. - PubMed
-
- Ljungman, M. & Zhang, F. (1996) Oncogene 13, 823-831. - PubMed
-
- McKay, B. C., Chen, F., Clarke, S. T., Wiggin, H. E., Harvey, L. M. & Ljungman, M. (2001) Mutat. Res. 485, 93-105. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

