The bchU gene of Chlorobium tepidum encodes the c-20 methyltransferase in bacteriochlorophyll c biosynthesis
- PMID: 15090495
- PMCID: PMC387796
- DOI: 10.1128/JB.186.9.2558-2566.2004
The bchU gene of Chlorobium tepidum encodes the c-20 methyltransferase in bacteriochlorophyll c biosynthesis
Abstract
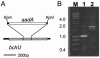
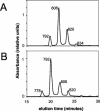
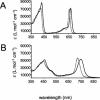
Bacteriochlorophylls (BChls) c and d, two of the major light-harvesting pigments in photosynthetic green sulfur bacteria, differ only by the presence of a methyl group at the C-20 methine bridge position in BChl c. A gene potentially encoding the C-20 methyltransferase, bchU, was identified by comparative analysis of the Chlorobium tepidum and Chloroflexus aurantiacus genome sequences. Homologs of this gene were amplified and sequenced from Chlorobium phaeobacteroides strain 1549, Chlorobium vibrioforme strain 8327d, and C. vibrioforme strain 8327c, which produce BChls e, d, and c, respectively. A single nucleotide insertion in the bchU gene of C. vibrioforme strain 8327d was found to cause a premature, in-frame stop codon and thus the formation of a truncated, nonfunctional gene product. The spontaneous mutant of this strain that produces BChl c (strain 8327c) has a second frameshift mutation that restores the correct reading frame in bchU. The bchU gene was inactivated in C. tepidum, a BChl c-producing species, and the resulting mutant produced only BChl d. Growth rate measurements showed that BChl c- and d-producing strains of the same organism (C. tepidum or C. vibrioforme) have similar growth rates at high and intermediate light intensities but that strains producing BChl c grow faster than those with BChl d at low light intensities. Thus, the bchU gene encodes the C-20 methyltransferase for BChl c biosynthesis in Chlorobium species, and methylation at the C-20 position to produce BChl c rather than BChl d confers a significant competitive advantage to green sulfur bacteria living at limiting red and near-infrared light intensities.
Figures





Comment in
-
Identification of a key step in the biosynthetic pathway of bacteriochlorophyll c and its implications for other known and unknown green sulfur bacteria.J Bacteriol. 2004 Aug;186(16):5187-8. doi: 10.1128/JB.186.16.5187-5188.2004. J Bacteriol. 2004. PMID: 15292118 Free PMC article. No abstract available.
References
-
- Armstrong, G. A., M. Alberti, F. Leach, and J. E. Hearst. 1989. Nucleotide sequence, organization, and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol. Gen. Genet. 216:254-268. - PubMed
-
- Armstrong, G. A., A. Schmidt, G. Sandmann, and J. E. Hearst. 1990. Genetic and biochemical characterization of carotenoid biosynthesis mutants of Rhodobacter capsulatus. J. Biol. Chem. 265:8329-8338. - PubMed
-
- Badenhop, F., S. Steiger, M. Sandmann, and G. Sandmann. 2003. Expression and biochemical characterization of the 1-HO-carotenoid methylase CrtF from Rhodobacter capsulatus. FEMS Microbiol. Lett. 222:237-242. - PubMed
-
- Bañeras, L., C. M. Borrego, and L. J. Garcia-Gil. 1999. Growth-rate-dependent bacteriochlorophyll c/d ratio in the antenna of Chlorobium limicola strain UdG6040. Arch. Microbiol. 171:350-354.
-
- Bickley, J., and R. J. Owen. 1995. Preparation of bacterial genomic DNA. Methods Mol. Biol. 46:141-147. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

