Conditional lethal amber mutations in essential Escherichia coli genes
- PMID: 15090508
- PMCID: PMC387789
- DOI: 10.1128/JB.186.9.2673-2681.2004
Conditional lethal amber mutations in essential Escherichia coli genes
Erratum in
- J Bacteriol. 2004 Dec;186(24):8547
Abstract
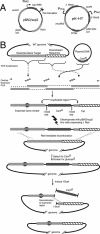
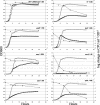
The essential genes of microorganisms encode biological functions important for survival and thus tend to be of high scientific interest. Drugs that interfere with essential functions are likely to be interesting candidates for antimicrobials. However, these genes are hard to study genetically because knockout mutations in them are by definition inviable. We recently described a conditional mutation system in Escherichia coli that uses a plasmid to produce an amber suppressor tRNA regulated by the arabinose promoter. This suppressor was used here in the construction of amber mutations in seven essential E. coli genes. Amber stop codons were introduced as "tagalong" mutations in the flanking DNA of a downstream antibiotic resistance marker by lambda red recombination. The drug marker was removed by expression of I-SceI meganuclease, leaving a markerless mutation. We demonstrate the method with the genes frr, gcpE, lpxC, map, murA, ppa, and rpsA. We were unable to isolate an amber mutation in ftsZ. Kinetics of cell death and morphological changes were measured following removal of arabinose. As expected given the wide range of cellular mechanisms represented, different mutants showed widely different death curves. All of the mutations were bactericidal except the mutation in gcpE, which was bacteriostatic. The strain carrying an amber mutation in murA was by far the most sensitive, showing rapid killing in nonpermissive medium. The MurA protein is critical for peptidoglycan synthesis and is the target for the antibiotic fosfomycin. Such experiments may inexpensively provide valuable information for the identification and prioritization of targets for antibiotic development.
Figures

References
-
- Arigoni, F., F. Talabot, M. Peitsch, M. D. Edgerton, E. Meldrum, E. Allet, R. Fish, T. Jamotte, M. L. Curchod, and H. Loferer. 1998. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16:851-856. - PubMed
-
- Armstrong, K. A., R. Acosta, E. Ledner, Y. Machida, M. Pancotto, M. McCormick, H. Ohtsubo, and E. Ohtsubo. 1984. A 37 × 103 molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J. Mol. Biol. 175:331-348. - PubMed
-
- Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. - PubMed
-
- Bockhorst, J., Y. Qiu, J. Glasner, M. Liu, F. Blattner, and M. Craven. 2003. Predicting bacterial transcription units using sequence and expression data. Bioinformatics 19(Suppl. 1):I34-I43. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

