The RNA-dependent RNA polymerase, QDE-1, is a rate-limiting factor in post-transcriptional gene silencing in Neurospora crassa
- PMID: 15090622
- PMCID: PMC407816
- DOI: 10.1093/nar/gkh530
The RNA-dependent RNA polymerase, QDE-1, is a rate-limiting factor in post-transcriptional gene silencing in Neurospora crassa
Abstract
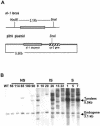
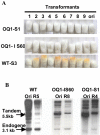
The RNA-dependent RNA polymerase (RdRP) qde-1 is an essential component of post-transcriptional gene silencing (PTGS), termed 'quelling' in the fungus Neurospora crassa. Here we show that the overexpression of QDE-1 results in a dramatic increase in the efficiency of quelling, with a concomitant net increase in the quantity of al-1 siRNAs. Moreover, in overexpressed strains there is a significant reduction in the number of transgenes required to induce quelling, and an increase in the phenotypic stability despite progressive loss of tandemly repeated transgenes, which normally determines reversion of a silenced phenotype to wild type. These data suggest that the activation and maintenance of silencing in Neurospora appear to rely both on the cellular amount of QDE-1 and the amount of transgenic copies producing RNA molecules that act as a substrate for the RdRP, implicating QDE-1 as a rate-limiting factor in PTGS.
Figures
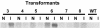


References
-
- Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. - PubMed
-
- Bernstein E., Caudy,A.C., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–336. - PubMed
-
- Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

