Mimotopes for alloreactive and conventional T cells in a peptide-MHC display library
- PMID: 15094798
- PMCID: PMC387264
- DOI: 10.1371/journal.pbio.0020090
Mimotopes for alloreactive and conventional T cells in a peptide-MHC display library
Abstract
The use of peptide libraries for the identification and characterization of T cell antigen peptide epitopes and mimotopes has been hampered by the need to form complexes between the peptides and an appropriate MHC molecule in order to construct a complete T cell ligand. We have developed a baculovirus-based peptide library method in which the sequence encoding the peptide is embedded within the genes for the MHC molecule in the viral DNA, such that insect cells infected with virus encoding a library of different peptides each displays a unique peptide-MHC complex on its surface. We have fished in such a library with two different fluorescent soluble T cell receptors (TCRs), one highly peptide specific and the other broadly allo-MHC specific and hypothesized to be much less focused on the peptide portion of the ligand. A single peptide sequence was selected by the former alphabetaTCR that, not unexpectedly, was highly related to the immunizing peptide. As hypothesized, the other alphabetaTCR selected a large family of peptides, related only by a similarity to the immunizing peptide at the p5 position. These findings have implications for the relative importance of peptide and MHC in TCR ligand recognition. This display method has broad applications in T cell epitope identification and manipulation and should be useful in general in studying interactions between complex proteins.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures


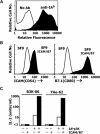
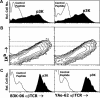
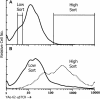


References
-
- Boder ET, Wittrup KD. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol. 2000;328:430–444. - PubMed
-
- Brophy SE, Holler PD, Kranz DM. A yeast display system for engineering functional peptide–MHC complexes. J Immunol Methods. 2003;272:235–246. - PubMed
-
- Crawford F, Kozono H, White J, Marrack P, Kappler J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 1998;8:675–682. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

