Operon prediction by comparative genomics: an application to the Synechococcus sp. WH8102 genome
- PMID: 15096577
- PMCID: PMC407844
- DOI: 10.1093/nar/gkh510
Operon prediction by comparative genomics: an application to the Synechococcus sp. WH8102 genome
Abstract
We present a computational method for operon prediction based on a comparative genomics approach. A group of consecutive genes is considered as a candidate operon if both their gene sequences and functions are conserved across several phylogenetically related genomes. In addition, various supporting data for operons are also collected through the application of public domain computer programs, and used in our prediction method. These include the prediction of conserved gene functions, promoter motifs and terminators. An apparent advantage of our approach over other operon prediction methods is that it does not require many experimental data (such as gene expression data and pathway data) as input. This feature makes it applicable to many newly sequenced genomes that do not have extensive experimental information. In order to validate our prediction, we have tested the method on Escherichia coli K12, in which operon structures have been extensively studied, through a comparative analysis against Haemophilus influenzae Rd and Salmonella typhimurium LT2. Our method successfully predicted most of the 237 known operons. After this initial validation, we then applied the method to a newly sequenced and annotated microbial genome, Synechococcus sp. WH8102, through a comparative genome analysis with two other cyanobacterial genomes, Prochlorococcus marinus sp. MED4 and P.marinus sp. MIT9313. Our results are consistent with previously reported results and statistics on operons in the literature.
Figures




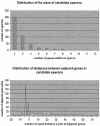

References
-
- Xu Y. (2004) Computational genome annotation. In Zhou,J., Thompson,D., Xu,Y. and Tidge,J. (eds), Microbial Functional Genomics. Wiley-LISS, Hoboken, NJ, pp. 41–66.
-
- Craven M., Page,D., Shavlik,J., Bockhorst,J. and Glasner,J. (2000) A probabilistic learning approach to whole-genome operon prediction. Proceedings of the 8th International Conference on Intelligent Systems for Molecular Biology, AAAI Press, San Diego, CA, pp. 116–127. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

